The hybrid history of zebrafish
- PMID: 39698833
- PMCID: PMC11797037
- DOI: 10.1093/g3journal/jkae299
The hybrid history of zebrafish
Abstract
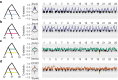
Since the description of zebrafish (Danio rerio) in 1822, the identity of its closest living relative has been unclear. To address this problem, we sequenced the exomes of 10 species in genus Danio, using the closely related Devario aequipinnatus as outgroup, to infer relationships across the 25 chromosomes of the zebrafish genome. The majority of relationships within Danio were remarkably consistent across all chromosomes. Relationships of chromosome segments, however, depended systematically upon their genomic location within zebrafish chromosomes. Regions near chromosome centers identified Danio kyathit and/or Danio aesculapii as the closest relative of zebrafish, while segments near chromosome ends supported only D. aesculapii as the zebrafish sister species. Genome-wide comparisons of derived character states revealed that danio relationships are inconsistent with a simple bifurcating species history but support an ancient hybrid origin of the D. rerio lineage by homoploid hybrid speciation. We also found evidence of more recent gene flow limited to the high recombination ends of chromosomes and several megabases of chromosome 20 with a history distinct from the rest of the genome. Additional insights gained from incorporating genome structure into a phylogenomic study demonstrate the utility of such an approach for future studies in other taxa. The multiple genomic histories of species in the genus Danio have important implications for comparative studies in these morphologically varied and beautiful species and for our understanding of the hybrid evolutionary history of zebrafish.
Keywords: danios; genome structure; hybrid species; introgression; phylogenomics; zebrafish.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The authors declare no conflicts of interest.
Figures





References
-
- Baldridge D, Wangler MF, Bowman AN, Yamamoto S; Undiagnosed Diseases Network; Schedl T, Pak SC, Postlethwait JH, Shin J, Solnica-Krezel L, et al. 2021. Model organisms contribute to diagnosis and discovery in the undiagnosed diseases network: current state and a future vision. Orphanet J Rare Dis. 16(1):206. doi:10.1186/s13023-021-01839-9. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
