Multiple Trait Bayesian Analysis of Partitioned Genetic Trends Accounting for Uncertainty in Genetic Parameters. An Example With the Pirenaica and Rubia Gallega Beef Cattle Breeds
- PMID: 39698940
- PMCID: PMC12149491
- DOI: 10.1111/jbg.12918
Multiple Trait Bayesian Analysis of Partitioned Genetic Trends Accounting for Uncertainty in Genetic Parameters. An Example With the Pirenaica and Rubia Gallega Beef Cattle Breeds
Abstract
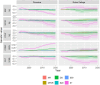
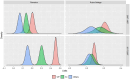
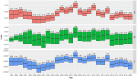
Genetic trends are a valuable tool for analysing the efficiency of breeding programs. They are calculated by averaging the predicted breeding values for all individuals born within a specific time period. Moreover, partitioned genetic trends allow dissecting the contributions of several selection paths to overall genetic progress. These trends are based on the linear relationship between breeding values and the Mendelian sampling terms of ancestors, enabling genetic trends to be split into contributions from different categories of individuals. However, (1) the use of predicted breeding values in calculating partitioned genetic trends depends on the variance components used and (2) a multiple trait analysis allows accounting for selection on correlated traits. These points are often not considered. To overcome these limitations, we present a software called "TM_TRENDS." This software performs a Bayesian analysis of partitioned genetic trends in a multiple trait model, accounting for uncertainty in the variance components. To illustrate the capabilities of this tool, we analysed the partitioned genetic trends for five traits (Birth Weight, Weight at 210 days, Cold Carcass Weight, Carcass Conformation, and Fatness Conformation) in two Spanish beef cattle breeds, Pirenaica and Rubia Gallega. The global genetic trends showed an increase in Carcass Conformation and a decrease in Birth Weight, Weight at 210 days, Cold Carcass Weight, and Fatness Conformation. These trends were partitioned into six categories: non-reproductive individuals, dams of females and non-reproductive individuals, dams of sires, sires with fewer than 20 progeny, sires between 20 and 50 progeny, and sires with more than 50 progeny. The results showed that the main source of genetic progress comes from sires with more than 50 progenies, followed by dams of males. Additionally, two additional features of the Bayesian analysis are presented: the calculation of the posterior probability of the global and partitioned genetic response between two time points, and the calculation of the posterior probability of positive (or negative) genetic progress.
© 2024 The Author(s). Journal of Animal Breeding and Genetics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Altarriba, J. , Yagüe G., Moreno C., and Varona L.. 2009. “Exploring the Possibilities of Genetic Improvement From Traceability Data. An Example in the Pirenaica Beef Cattle.” Livestock Science 125: 115–120. 10.1016/j.livsci.2009.03.013. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

