Personalized deep learning auto-segmentation models for adaptive fractionated magnetic resonance-guided radiation therapy of the abdomen
- PMID: 39699250
- PMCID: PMC11972049
- DOI: 10.1002/mp.17580
Personalized deep learning auto-segmentation models for adaptive fractionated magnetic resonance-guided radiation therapy of the abdomen
Abstract
Background: Manual contour corrections during fractionated magnetic resonance (MR)-guided radiotherapy (MRgRT) are time-consuming. Conventional population models for deep learning auto-segmentation might be suboptimal for MRgRT at MR-Linacs since they do not incorporate manual segmentation from treatment planning and previous fractions.
Purpose: In this work, we investigate patient-specific (PS) auto-segmentation methods leveraging expert-segmented planning and prior fraction MR images (MRIs) to improve auto-segmentation on consecutive treatment days.
Materials and methods: Data from 151 abdominal cancer patients treated at a 0.35 T MR-Linac (151 planning and 215 fraction MRIs) were included. Population baseline models (BMs) were trained on 107 planning MRIs for one-class segmentation of the aorta, bowel, duodenum, kidneys, liver, spinal canal, and stomach. PS models were obtained by fine-tuning the BMs using the planning MRI ( ). Maximal improvement by continuously updating the PS models was investigated by adding the first four out of five fraction MRIs ( ). Similarly, PS models without BM were trained ( and ). All hyperparameters were optimized using 23 patients, and the methods were tested on the remaining 21 patients. Evaluation involved Dice similarity coefficient (DSC), average ( ) and the 95th percentile (HD95) Hausdorff distance. A qualitative contour assessment by a radiation oncologist was performed for BM, , and .
Results: and networks had the best geometric performance. and BMs showed similar DSC and HDs values, however models outperformed BMs. predictions scored the best in the qualitative evaluation, followed by the BMs and models.
Conclusion: Personalized auto-segmentation models outperformed the population BMs. In most cases, delineations were judged to be directly usable for treatment adaptation without further corrections, suggesting a potential time saving during fractionated treatment.
Keywords: MR‐Linac; auto‐segmentation; patient‐specific transfer learning.
© 2024 The Author(s). Medical Physics published by Wiley Periodicals LLC on behalf of American Association of Physicists in Medicine.
Conflict of interest statement
The Department of Radiation Oncology of the University Hospital of LMU Munich has research agreements with Elekta and Brainlab.
Figures

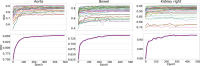

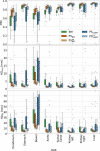
 percentile (), and average () Hausdorff distance for the 5th fractions of the 21 test patients. For all organs‐at‐risk the performance of the following models are compared: the BMs, PS models generated by fine‐tuning the BMs with one () and five MRIs (), as well as PS models trained from scratch with one () and with 5 MRIs () of a given patient. BMs, baseline models; HD, Hausdorff distance; MRI, magnetic resonance imaging; PS, patient‐specific.
percentile (), and average () Hausdorff distance for the 5th fractions of the 21 test patients. For all organs‐at‐risk the performance of the following models are compared: the BMs, PS models generated by fine‐tuning the BMs with one () and five MRIs (), as well as PS models trained from scratch with one () and with 5 MRIs () of a given patient. BMs, baseline models; HD, Hausdorff distance; MRI, magnetic resonance imaging; PS, patient‐specific.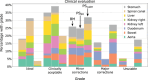
References
-
- Tijssen RH, Philippens ME, Paulson ES, et al. MRI commissioning of 1.5 T MR‐linac systems–a multi‐institutional study. Radiother Oncol. 2019;132:114‐120. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

