Detection of CRISPR‒Cas and type I R-M systems in Klebsiella pneumoniae of human and animal origins and their relationship to antibiotic resistance and virulence
- PMID: 39699265
- PMCID: PMC11792477
- DOI: 10.1128/spectrum.00009-24
Detection of CRISPR‒Cas and type I R-M systems in Klebsiella pneumoniae of human and animal origins and their relationship to antibiotic resistance and virulence
Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR)‒CRISPR-associated protein (Cas) and restriction‒modification (R-M) systems are important immune systems in bacteria. Information about the distributions of these two systems in Klebsiella pneumoniae from different hosts and their mutual effect on antibiotic resistance and virulence is still limited. In this study, the whole genomes of 520 strains of K. pneumoniae from GenBank, including 325 from humans and 195 from animals, were collected for CRISPR‒Cas systems and type I R-M systems, virulence genes, antibiotic resistance genes, and multilocus sequence typing detection. The results showed that host origin had no obvious influence on the distributions of the two systems (CRISPR‒Cas systems in 29.8% and 24.1%, type I R-M systems in 9.8% and 11.8% of human-origin and animal-origin strains, respectively) in K. pneumoniae. Identical spacer sequences from different hosts demonstrated there was a risk of human-animal transmission. All virulence genes (yersiniabactin, colibactin, aerobactin, salmochelin, rmpADC, and rmpA2) detection rates were higher when only the CRISPR‒Cas systems were present but were all reduced when coexisting with type I R-M systems. However, a lower prevalence of most antibiotic-resistance genes was found when the CRISPR‒Cas systems were alone, and when type I R-M systems were coexisting, some of the antibiotic resistance gene incidence rates were even lower (quinolones, macrolides, tetracyclines and carbapenems), and some of them were higher instead (aminoglycosides, clindamycins, rifampicin-associated, sulfonamides, methotrexates, beta-lactamases and ultrabroad-spectrum beta-lactamases). The synergistic and opposed effects of the two systems on virulence and antibiotic-resistance genes need further study.IMPORTANCEK. pneumoniae is an important opportunistic pathogen responsible for both human and animal infections, and the emergence of hypervirulent and multidrug-resistant K. pneumoniae has made it difficult to control this pathogen worldwide. Here, we find that CRISPR‒Cas and restriction-modification systems, which function as adaptive and innate immune systems in bacteria, have synergistic and opposed effects on virulence and antibiotic resistance genes in K. pneumoniae. Moreover, this study provides insights into the distributions of the two systems in K. pneumoniae from different hosts, and there is no significant difference in the prevalence of the two systems among K. pneumoniae spp. In addition, this study also characterizes the CRISPR arrays of K. pneumoniae from different hosts, suggesting that the strains sharing the same spacer sequences have the potential to spread between humans and animals.
Keywords: CRISPR‒Cas system; Klebsiella pneumoniae; R–M system; antibiotic resistance; multilocus sequence typing; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

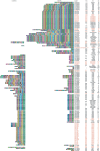
References
-
- Zhou Y, Tang Y, Fu P, Tian D, Yu L, Huang Y, Li G, Li M, Wang Y, Yang Z, Xu X, Yin Z, Zhou D, Poirel L, Jiang X. 2020. The type I-E CRISPR-Cas system influences the acquisition of bla(KPC)-IncF plasmid in Klebsiella pneumonia. Emerg Microbes Infect 9:1011–1022. doi: 10.1080/22221751.2020.1763209 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

