Single-molecule studies of repair proteins in base excision repair
- PMID: 39701025
- PMCID: PMC11788526
- DOI: 10.5483/BMBRep.2024-0178
Single-molecule studies of repair proteins in base excision repair
Abstract
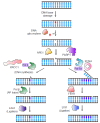
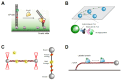
Base excision repair (BER) is an essential cellular mechanism that repairs small, non-helix-distorting base lesions in DNA, resulting from oxidative damage, alkylation, deamination, or hydrolysis. This review highlights recent advances in understanding the molecular mechanisms of BER enzymes through single-molecule studies. We discuss the roles of DNA glycosylases in lesion recognition and excision, with a focus on facilitated diffusion mechanisms such as sliding and hopping that enable efficient genome scanning. The dynamics of apurinic/apyrimidinic endonucleases, especially the coordination between APE1 and DNA polymerase β (Pol β), are explored to demonstrate their crucial roles in processing abasic sites. The review further explores the short-patch and long-patch BER pathways, emphasizing the activities of Pol β, XRCC1, PARP1, FEN1, and PCNA in supporting repair synthesis and ligation. Additionally, we highlight the emerging role of UV-DDB as a general damage sensor in BER, extending its recognized function beyond nucleotide excision repair. Single-molecule techniques have been instrumental in uncovering the complex interactions and mechanisms of BER proteins, offering unprecedented insights that could guide future therapeutic strategies for maintaining genomic stability. [BMB Reports 2025; 58(1): 17-23].
Conflict of interest statement
The authors have no conflicting interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

