Single-molecule DNA-flow stretching assay as a versatile hybrid tool for investigating DNA-protein interactions
- PMID: 39701027
- PMCID: PMC11788529
- DOI: 10.5483/BMBRep.2024-0177
Single-molecule DNA-flow stretching assay as a versatile hybrid tool for investigating DNA-protein interactions
Abstract
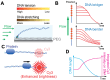
Single-molecule techniques allow researchers to investigate individual molecules and obtain unprecedented details of the heterogeneous nature of biological entities. They play instrumental roles in studying DNA-protein interactions due to the ability to visualize DNA or proteins and to manipulate individual DNA molecules by applying force or torque. Here, we describe single-molecule DNA-flow stretching assays as hybrid tools that combine forces with fluorescence. We also review how widely these assays are utilized in elucidating working mechanisms of DNA-binding proteins. Additionally, we provide a brief explanation of various efforts to prepare DNA substrates with desired internal protein-binding sequences. More complicated needs for DNA-protein interaction research have led to improvements in single-molecule DNA flow-stretching techniques. Several DNA flow-stretching variants such as DNA curtain, DNA motion capture assays, and protein-induced fluorescence enhancement (PIFE) are introduced in this mini review. Singlemolecule DNA flow-stretching assays will keep contributing to our understanding of how DNA-binding proteins function due to their multiplexed, versatile, and robust capabilities. [BMB Reports 2025; 58(1): 41-51].
Conflict of interest statement
The authors have no conflicting interests.
Figures


Similar articles
-
Initial state of DNA-Dye complex sets the stage for protein induced fluorescence modulation.Nat Commun. 2019 May 8;10(1):2104. doi: 10.1038/s41467-019-10137-9. Nat Commun. 2019. PMID: 31068591 Free PMC article.
-
Multiplexed single-molecule flow-stretching bead assay for DNA enzymology.BMB Rep. 2019 Oct;52(10):589-594. doi: 10.5483/BMBRep.2019.52.10.170. BMB Rep. 2019. PMID: 31401983 Free PMC article. Review.
-
Observing Bacterial Chromatin Protein-DNA Interactions by Combining DNA Flow-Stretching with Single-Molecule Imaging.Methods Mol Biol. 2018;1837:277-299. doi: 10.1007/978-1-4939-8675-0_15. Methods Mol Biol. 2018. PMID: 30109616
-
When Force Met Fluorescence: Single-Molecule Manipulation and Visualization of Protein-DNA Interactions.Annu Rev Biophys. 2024 Jul;53(1):169-191. doi: 10.1146/annurev-biophys-030822-032904. Epub 2024 Jun 28. Annu Rev Biophys. 2024. PMID: 38237015 Review.
-
Shining a Spotlight on DNA: Single-Molecule Methods to Visualise DNA.Molecules. 2019 Jan 30;24(3):491. doi: 10.3390/molecules24030491. Molecules. 2019. PMID: 30704053 Free PMC article. Review.
Cited by
-
Probing the effect of PEG-DNA interactions and buffer viscosity on tethered DNA in shear flow.PLoS One. 2025 Aug 25;20(8):e0329961. doi: 10.1371/journal.pone.0329961. eCollection 2025. PLoS One. 2025. PMID: 40853970 Free PMC article.
-
ParB C-terminal lysine residues are essential for dimerization, in vitro DNA sliding and in vivo function.bioRxiv [Preprint]. 2025 Jun 15:2025.06.10.659001. doi: 10.1101/2025.06.10.659001. bioRxiv. 2025. PMID: 40661362 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

