Higher order interaction analysis quantifies coordination in the epigenome revealing novel biological relationships in Kabuki syndrome
- PMID: 39701600
- PMCID: PMC11658816
- DOI: 10.1093/bib/bbae667
Higher order interaction analysis quantifies coordination in the epigenome revealing novel biological relationships in Kabuki syndrome
Abstract
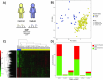
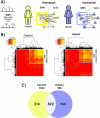
Complex direct and indirect relationships between multiple variables, termed higher order interactions (HOIs), are characteristics of all natural systems. Traditional differential and network analyses fail to account for the omic datasets richness and miss HOIs. We investigated peripheral blood DNA methylation data from Kabuki syndrome type 1 (KS1) and control individuals, identified 2,002 differentially methylated points (DMPs), and inferred 17 differentially methylated regions, which represent only 189 DMPs. We applied hypergraph models to measure HOIs on all the CpGs and revealed differences in the coordination of DMPs with lower entropy and higher coordination of the peripheral epigenome in KS1 implying reduced network complexity. Hypergraphs also capture epigenomic trans-relationships, and identify biologically relevant pathways that escape the standard analyses. These findings construct the basis of a suitable model for the analysis of organization in the epigenome in rare diseases, which can be applied to investigate mechanism in big data.
Keywords: DNA methylation; KMT2D; Kabuki syndrome; integration analysis.
© The Author(s) 2024. Published by Oxford University Press.
Figures





References
-
- Battiston F, Cencetti G, Iacopini I. et al. . Networks beyond pairwise interactions: structure and dynamics. Phys Rep 2020;874:1–92. 10.1016/j.physrep.2020.05.004. - DOI
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

