Prediction of miRNA-disease associations based on PCA and cascade forest
- PMID: 39701957
- PMCID: PMC11660965
- DOI: 10.1186/s12859-024-05999-w
Prediction of miRNA-disease associations based on PCA and cascade forest
Abstract
Background: As a key non-coding RNA molecule, miRNA profoundly affects gene expression regulation and connects to the pathological processes of several kinds of human diseases. However, conventional experimental methods for validating miRNA-disease associations are laborious. Consequently, the development of efficient and reliable computational prediction models is crucial for the identification and validation of these associations.
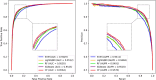
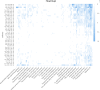
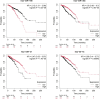
Results: In this research, we developed the PCACFMDA method to predict the potential associations between miRNAs and diseases. To construct a multidimensional feature matrix, we consider the fusion similarities of miRNA and disease and miRNA-disease pairs. We then use principal component analysis(PCA) to reduce data complexity and extract low-dimensional features. Subsequently, a tuned cascade forest is used to mine the features and output prediction scores deeply. The results of the 5-fold cross-validation using the HMDD v2.0 database indicate that the PCACFMDA algorithm achieved an AUC of 98.56%. Additionally, we perform case studies on breast, esophageal and lung neoplasms. The findings revealed that the top 50 miRNAs most strongly linked to each disease have been validated.
Conclusions: Based on PCA and optimized cascade forests, we propose the PCACFMDA model for predicting undiscovered miRNA-disease associations. The experimental results demonstrate superior prediction performance and commendable stability. Consequently, the PCACFMDA is a potent instrument for in-depth exploration of miRNA-disease associations.
Keywords: Cascade forest; Ensemble learning; PCA; miRNA-disease Association.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interest.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

