Environmental sex reversal in parrotfish does not cause differences in the structure of their gut microbial communities
- PMID: 39701987
- PMCID: PMC11657377
- DOI: 10.1186/s12866-024-03698-3
Environmental sex reversal in parrotfish does not cause differences in the structure of their gut microbial communities
Abstract
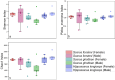
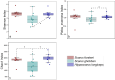
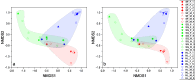
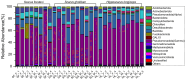
Parrotfish are a common fish in coral reef areas, but little is known about their gut microbial communities. In addition, parrotfish are capable of sex reversal, usually some males are sexually reversed from females, and it is still not known whether this sex reversal leads to significant changes in gut microbial communities. In this study, we investigated the gut microbial communities of three species of parrotfish including Scarus forsteni (4 females and 4 sex-reversed males), Scarus ghobban (5 females and 5 sex-reversed males), and Hipposcarus longiceps (5 females and 5 sex-reversed males) by using high-throughput sequencing technology. The gut microbial communities of these three species were mainly composed of Pseudomonadota (class Gammaproteobacteria) and Bacillota, while at the family level, they mainly included Vibrionaceae, Burkholderiaceae, Enterobacteriaceae, Streptococcacea, and Erwiniaceae. Although at the genus level, there were a large number of unclassified lineages, the remaining gut microorganisms were mainly composed of Vibrio, Photobacterium, Enterococcus and Lactococcus. Furthermore, we did not find significant differences in gut microbial community structure between the female parrotfish and corresponding female reversed males within each species, even in terms of the structure of gut microbial functional information obtained from 16 S rRNA gene sequence predictions. However, the gut microbial communities of these three species of parrotfish differed significantly not only in their community structure but also in their microbial functional information structure, mainly in terms of aspartate and asparagine biosynthesis, histidine degradation, inositol degradation, heptose biosynthesis, chitin derivatives degradation, enterobactin biosynthesis, and thiazole biosynthesis. Our study provides essential gut microbial community data for understanding the physiology and sex reversal phenomenon in parrotfish.
Keywords: 16S rRNA gene sequence; Gut microflora; Microbial communities; Parrotfish; Sex reversal.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All fish handling procedures were approved by the Animal Ethics Committee, School of Life Science and Technology, Wuhan Polytechnic University (Authorization No.: WPU202204008). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208(3–4):191–364. - DOI
MeSH terms
Substances
Grants and funding
- 32102769/National Natural Science Foundation of China
- 32102769/National Natural Science Foundation of China
- 32102769/National Natural Science Foundation of China
- 32102769/National Natural Science Foundation of China
- 2022CFB403/Natural Science Foundation of Hubei Province of China
- 2022CFB403/Natural Science Foundation of Hubei Province of China
- 2022CFB403/Natural Science Foundation of Hubei Province of China
- Q20221614/Scientific Research Items Foundation of Hubei Educational Committee
- Q20221614/Scientific Research Items Foundation of Hubei Educational Committee
- Q20221614/Scientific Research Items Foundation of Hubei Educational Committee
LinkOut - more resources
Full Text Sources

