HGVS Nomenclature 2024: improvements to community engagement, usability, and computability
- PMID: 39702242
- PMCID: PMC11660784
- DOI: 10.1186/s13073-024-01421-5
HGVS Nomenclature 2024: improvements to community engagement, usability, and computability
Abstract
Background: The Human Genome Variation Society (HGVS) Nomenclature is the global standard for describing and communicating variants in DNA, RNA, and protein sequences in clinical and research genomics. This manuscript details recent updates to the HGVS Nomenclature, highlighting improvements in governance, community engagement, website functionality, and underlying implementation of the standard.
Methods: The HGVS Variant Nomenclature Committee (HVNC) now operates under the Human Genome Organization (HUGO), facilitating broader community feedback and collaboration with related standards organizations. The website has been redesigned using modern documentation tools and practices. The specification was updated to include guidance for transcript selection and to align with recent cross-consortia recommendations for the representation of gene fusions. A formal computational grammar was introduced to improve the precision and consistency of variant descriptions.
Results: Major improvements in HGVS Nomenclature v. 21.1 include a redesigned website with enhanced navigation, search functionality, and mobile responsiveness; a new versioning policy aligned with software management practices; formal mechanisms for community feedback and change proposals; and adoption of Extended Backus-Naur Form (EBNF) for defining syntax. The specification now recommends MANE Select transcripts where appropriate and includes updated guidance for representing adjoined transcripts and gene fusions. All content is freely available under permissive licenses at hgvs-nomenclature.org.
Conclusions: These advancements establish a more sustainable foundation for maintaining and evolving the HGVS Nomenclature while improving its accessibility and utility. The introduction of formal computational grammar marks a crucial step toward unambiguous variant descriptions that can be reliably processed by both humans and machines. Combined with enhanced community engagement mechanisms and improved guidance, these changes position the HGVS Nomenclature to better serve the evolving needs of clinical and research genomics while maintaining the stability that users require.
Keywords: Bioinformatics; Clinical genomics; HGVS; Sequence variation; Standards.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: RH is an employee of GenQA. RKH is an employee and shareholder of MyOme, Inc. RT is an employee of EMQN CIC.
Figures


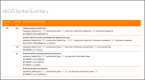
References
-
- den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016. Available from: 10.1002/humu.22981. - PubMed
-
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11479744. - DOI - PubMed
-
- McGowan-Jordan J, Moore S, Hastings R, editors. ISCN 2020: an international system for human cytogenomic nomenclature. Cytogenet Genome Res. 2020;160:341–503. Available from: https://iscn.karger.com/. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

