Histone variant H3.5 in testicular cell differentiation and its interactions with histone chaperones
- PMID: 39702777
- PMCID: PMC11659419
- DOI: 10.1038/s41598-024-83206-9
Histone variant H3.5 in testicular cell differentiation and its interactions with histone chaperones
Abstract
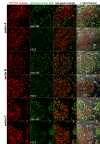
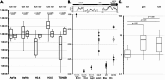
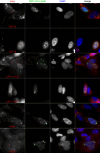
Testicular cell differentiation is a highly regulated process, essential for male reproductive health. The histone variant H3.5 is apparently a critical player in this intricate orchestra of cell types, but its regulation and function remains poorly understood. To elucidate its role, we fractionized testicular cells using c-Kit/CD117 as a separation marker and analyzed H3.5 expression. Further, we investigated the regulation of H3.5 expression using public data repositories. We explored DNA methylation patterns in specific regions of the H3-5 gene and assessed H3-5 copy number gain in seminoma specimens. Additionally, we examined the testicular localization of H3.5 and its histone chaperone interactions to understand its regulation at the protein level. We used qRT-PCR, MeDIP, and qPCR to study H3.5 expression and DNA methylation in various cell types. H3-5 copy number gain was analyzed using qPCR. Protein interactions were investigated through fluorescence-2-hybrid assays in baby hamster kidney cells. H3.5 is primarily enriched in spermatocytes. DNA methylation of a CpG island overlapping the H3-5 promoter appeared to be involved in the tissue-specific regulation of H3.5 expression. Elevated H3.5 expression was observed in seminoma specimens, suggesting a potential link to testicular tumors. H3-5 copy number gain was associated with elevated H3.5 expression in seminoma specimens. Furthermore, we identified physical interactions between H3.5 and histone chaperones Asf1a and Asf1b, HIRA, CAF p150 and DAXX, shedding light on the protein-level regulation of H3.5. These findings provide valuable insights into the molecular mechanisms governing testicular cell differentiation and the potential role of H3.5 in testicular pathologies.
Keywords: Chromatin remodeling; Epigenetic Plasticity; Epigenetic regulation; Histone chaperones; Histone variants; Spermatogenesis.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval: This study was conducted with approval of the Witten/Herdecke University Ethics board (No’s. 90/2011 and 138/2013). In addition, for investigations involving human subjects, informed consent has been obtained from all participants. Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research. Research involving human research participants were performed in accordance with the Declaration of Helsinki. Informed consent: Informed written consent was obtained by all involved donors.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

