Bleximenib, the novel menin-KMT2A inhibitor JNJ-75276617, impairs long-term proliferation and immune evasion in acute myeloid leukemia
- PMID: 39704147
- PMCID: PMC12130779
- DOI: 10.3324/haematol.2024.285616
Bleximenib, the novel menin-KMT2A inhibitor JNJ-75276617, impairs long-term proliferation and immune evasion in acute myeloid leukemia
Abstract
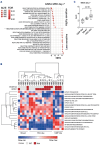
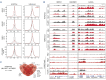
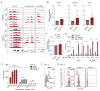
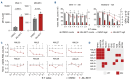
Acute myeloid leukemia (AML) remains challenging to treat, which in part relates to genetic heterogeneity of the disease, to the protective tumor microenvironment driving resistance to therapy, and also to immune evasion characteristics of leukemic cells. Targeting epigenetic programs in AML provides an attractive opportunity to impair long-term proliferation and induce differentiation. The novel inhibitor JNJ-75276617 (bleximenib) targets the menin-KMT2A interaction and has shown preclinical efficacy in AML.1 Here, we provide mechanistic insights into how JNJ-75276617 impairs proliferation and drives differentiation of primary AML patient cells. A large-scale drug screen was set up in which genetic alterations and quantitative proteomics were compared with drug sensitivity in a preclinical setting, which revealed that granulocyte-macrophage progenitor (GMP)-like AML display the greatest sensitivity. Furthermore, we identified that NPM1c/DNMT3Amut AML are sensitive, and some NPM1wt AML subtypes without KMT2A-MLLT3 rearrangements benefit from menin-KMT2A inhibition. Genome-wide chromatin immunoprecipitation- sequencing studies revealed patient-specific epigenetic alterations upon JNJ-75276617 treatment, uncovering a striking upregulation of MHC class I and class II expression as a consequence of epigenetic changes upon menin-KMT2A inhibition, independent of MEIS1 loss but involving CIITA activation. Functionally, this results in enhanced sensitivity of leukemic blasts to T-cell-mediated cytotoxicity in allogeneic and autologous settings. Our data indicate that JNJ-75276617 provides a potential therapeutic approach whereby not only proliferation is impaired and differentiation is induced, but whereby therapeutic benefit might also be achieved by reactivating the antigen presentation machinery.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

