MRE11-independent effects of Mirin on mitochondrial DNA integrity and cellular immune responses
- PMID: 39705374
- PMCID: PMC11809308
- DOI: 10.1091/mbc.E24-01-0002
MRE11-independent effects of Mirin on mitochondrial DNA integrity and cellular immune responses
Abstract
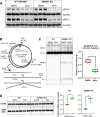
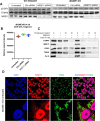
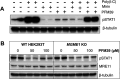
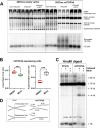
Mirin, a chemical inhibitor of MRE11, has been recently reported to suppress immune response triggered by mitochondrial DNA (mtDNA) breakage and release during replication stalling. We show that while Mirin reduces mitochondrial replication fork breakage in mitochondrial 3´-exonuclease MGME1 deficient cells, this effect occurs independently of MRE11. We also discovered that Mirin directly inhibits cellular immune responses, as shown by its suppression of STAT1 phosphorylation in Poly (I:C)-treated cells. Furthermore, Mirin also altered mtDNA supercoiling and accumulation of hemicatenated replication termination intermediates-hallmarks of topoisomerase dysfunction-while mitigating topological changes induced by the overexpression of mitochondrial TOP3A, including TOP3A-dependent strand breakage at the noncoding region of mtDNA. Although Mirin does not seem to inhibit TOP3A activity in vitro, our findings demonstrate its MRE11-independent effects in cells and give insight into the mechanisms of the maintenance of mtDNA integrity.
Conflict of interest statement
Conflicts of Interests: The authors declare no financial conflict of interest.
Figures


References
-
- Bizard AH, Allemand JF, Hassenkam T, Paramasivam M, Sarlos K, Singh MI, Hickson ID (2019). PICH and TOP3A cooperate to induce positive DNA supercoiling. Nat Struct Mol Biol 26, 267–274. - PubMed
-
- Cookson JC, Dai F, Smith V, Heald RA, Laughton CA, Stevens MF, Burger AM (2005). Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate (RHPS4) in vitro: activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents. Mol Pharmacol 68, 1551–1558. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

