A Novel Deer Antler-Inspired Bone Graft Triggers Rapid Bone Regeneration
- PMID: 39707695
- PMCID: PMC11817900
- DOI: 10.1002/adma.202411571
A Novel Deer Antler-Inspired Bone Graft Triggers Rapid Bone Regeneration
Abstract
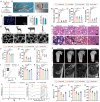
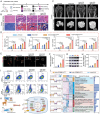
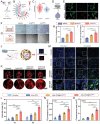
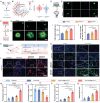
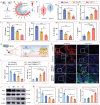
Adult mammals are unable to regenerate bulky bone tissues, making large bone defects clinically challenging. Deer antler represents an exception to this rule, exhibiting the fastest bony growth in mammals, offering a unique opportunity to explore novel strategies for rapid bone regeneration. Here, a bone graft exploiting the biochemical, biophysical, and structural characteristics of antlers is constructed. It is decellularized antler cancellous bone (antler-DCB) to obtain a bone scaffold. Then, an antler-based bone graft is constructed by integrating antler-DCB with antler-derived biological signals, delivered by extracellular vesicles (EVs) from antler blastema progenitor cells (ABPCs), a novel stem cells responsible for antlerogenesis is discovered. The antler-based bone graft transformed bone marrow stromal cells into cells with an ABPC-like phenotype and transcriptomic signature. In vivo, the antler-based graft triggered rapid bone formation in a rat model, with doubled volume of newly formed bones than commercial DCBs. In addition, the antler-based graft orchestrated a coordinated process of vascularization, neurogenesis, and immunomodulation during osteogenesis, partially imitating early antlerogenesis. These findings provide practical insights to develop a therapeutic intervention for treating severe bone defects.
Keywords: angiogenesis; antler‐based bone graft; bioinspiration; bone regeneration; neurogenesis.
© 2024 The Author(s). Advanced Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- a) Valtanen R. S., Yang Y. P., Gurtner G. C., Maloney W. J., Lowenberg D. W., Injury 2021, 52, S72; - PubMed
- b) Park W. B., Park J. S., Han J. Y., Kang P., Medicina (Kaunas) 2022, 58; - PMC - PubMed
- c) Nefjodovs V., Andze L., Andzs M., Filipova I., Tupciauskas R., Vecbiskena L., Kapickis M., J. Funct. Biomater. 2023, 14, 266. - PMC - PubMed
-
- Zhang H., Zhang M., Zhai D., Qin C., Wang Y., Ma J., Zhuang H., Shi Z., Wang L., Wu C., Adv. Mater. 2023, 35, 2302716. - PubMed
-
- Stegen S., Carmeliet G., Bone 2018, 115, 50. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

