MiCK: a database of gut microbial genes linked with chemoresistance in cancer patients
- PMID: 39707929
- PMCID: PMC11662283
- DOI: 10.1093/database/baae124
MiCK: a database of gut microbial genes linked with chemoresistance in cancer patients
Abstract
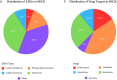
Cancer remains a global health challenge, with significant morbidity and mortality rates. In 2020, cancer caused nearly 10 million deaths, making it the second leading cause of death worldwide. The emergence of chemoresistance has become a major hurdle in successfully treating cancer patients. Recently, human gut microbes have been recognized for their role in modulating drug efficacy through their metabolites, ultimately leading to chemoresistance. The currently available databases are limited to knowledge regarding the interactions between gut microbiome and drugs. However, a database containing the human gut microbial gene sequences, and their effect on the efficacy of chemotherapy for cancer patients has not yet been developed. To address this challenge, we present the Microbial Chemoresistance Knowledgebase (MiCK), a comprehensive database that catalogs microbial gene sequences associated with chemoresistance. MiCK contains 1.6 million sequences of 29 gene types linked to chemoresistance and drug metabolism, curated manually from recent literature and sequence databases. The database can support downstream analysis as it provides a user-friendly web interface for sequence search and download functionalities. MiCK aims to facilitate the understanding and mitigation of chemoresistance in cancers by serving as a valuable resource for researchers. Database URL: https://microbialchemreskb.com/.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

Similar articles
-
gutMGene: a comprehensive database for target genes of gut microbes and microbial metabolites.Nucleic Acids Res. 2022 Jan 7;50(D1):D795-D800. doi: 10.1093/nar/gkab786. Nucleic Acids Res. 2022. PMID: 34500458 Free PMC article.
-
GMMAD: a comprehensive database of human gut microbial metabolite associations with diseases.BMC Genomics. 2023 Aug 24;24(1):482. doi: 10.1186/s12864-023-09599-5. BMC Genomics. 2023. PMID: 37620754 Free PMC article.
-
gutMDisorder: a comprehensive database for dysbiosis of the gut microbiota in disorders and interventions.Nucleic Acids Res. 2020 Jan 8;48(D1):D554-D560. doi: 10.1093/nar/gkz843. Nucleic Acids Res. 2020. PMID: 31584099 Free PMC article.
-
Interplay of gut microbiome, fatty acids, and the endocannabinoid system in regulating development, progression, immunomodulation, and chemoresistance of cancer.Nutrition. 2022 Nov-Dec;103-104:111787. doi: 10.1016/j.nut.2022.111787. Epub 2022 Jul 16. Nutrition. 2022. PMID: 36055123 Review.
-
What "The Cancer Genome Atlas" database tells us about the role of ATP-binding cassette (ABC) proteins in chemoresistance to anticancer drugs.Expert Opin Drug Metab Toxicol. 2019 Jul;15(7):577-593. doi: 10.1080/17425255.2019.1631285. Epub 2019 Jun 23. Expert Opin Drug Metab Toxicol. 2019. PMID: 31185182 Review.
References
-
- WHO. Cancer Today. 2022. https://gco.iarc.fr/today/en (3 April 2024, date last accessed).
-
- WHO. Cancer. 2022. https://www.who.int/news-room/fact-sheets/detail/cancer (2 July 2024, date last accessed).
-
- Elgemeie GH, Mohamed-Ezzat RA. Pyrimidine-based Anticancer Drugs, New Strategies Targeting Cancer Metabolism 1st edn. Radarweg 29, 1043 NX Amsterdam, The Netherlands: Elsevier, 2022, 107–42.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

