Parallel Proteomic and Transcriptomic Microenvironment Mapping (μMap) of Nuclear Condensates in Living Cells
- PMID: 39707993
- PMCID: PMC11792175
- DOI: 10.1021/jacs.4c11612
Parallel Proteomic and Transcriptomic Microenvironment Mapping (μMap) of Nuclear Condensates in Living Cells
Abstract
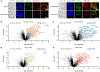
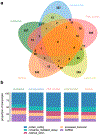
Cellular activity is spatially organized across different organelles. While several structures are well-characterized, many organelles have unknown roles. Profiling biomolecular composition is key to understanding function but is difficult to achieve in the context of small, dynamic structures. Photoproximity labeling has emerged as a powerful tool for mapping these interaction networks, yet maximizing catalyst localization and reducing toxicity remains challenging in live cell applications. Here, we disclose a new intracellular photocatalyst with minimal cytotoxicity and off-target binding, and we utilize this catalyst for HaloTag-based microenvironment-mapping (μMap) to spatially catalog subnuclear condensates in living cells. We also specifically develop a novel RNA-focused workflow (μMap-seq) to enable parallel transcriptomic and proteomic profiling of these structures. After validating the accuracy of our approach, we generate a spatial map across the nucleolus, nuclear lamina, Cajal bodies, paraspeckles, and PML bodies. These results provide potential new insights into RNA metabolism and gene regulation while significantly expanding the μMap platform for improved live-cell proximity labeling in biological systems.
Conflict of interest statement
The authors declare the following competing financial interest(s): DWCM declares an ownership interest in the company Dexterity Pharma LLC, which has commercialized materi-als used in this work.
Figures




References
-
- Spannl S; Tereshchenko M; Mastromarco GJ; Ihn SJ; Lee HO Biomolecular condensates in neurodegeneration and cancer. Traffic 2019, 20 (12), 890–911. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

