Comparison of whole genome sequencing typing tools for the typing of Belgian Legionella pneumophila outbreaks isolates
- PMID: 39708273
- PMCID: PMC11880184
- DOI: 10.1007/s10096-024-05013-4
Comparison of whole genome sequencing typing tools for the typing of Belgian Legionella pneumophila outbreaks isolates
Abstract
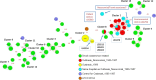
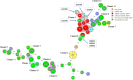
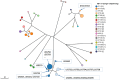
Whole genome sequencing (WGS) marks a turning point for outbreak investigations for microorganisms related to public health matters, like Legionella pneumophila (Lp). Here, we evaluated the available Lp WGS typing tools for isolates of previously documented Belgian outbreaks, as well as small groups of related and non-related isolates. One reference strain and 77 clinical and environmental isolates were evaluated. Seven isolates belong to a Sequence Type (ST) 36 outbreak in 1999 and sixteen (ten clinical, two matching environmental and four non-related controls) belong to another ST1 outbreak in 1985-1987. The remaining isolates belong to small groups of related and non-related isolates of diverse ST's. WGS was performed and data were analysed using whole genome (wg) and core genome (cg) multilocus sequence typing (MLST) with "Ridom SeqSphere + " (cgMLST), "Applied Maths-Bionumerics" (wgMLST) and the 50 loci cgMLST (CDC/ESGLI_ESCMID). Results of the three tools were concordant with the traditional Sequence Based Typing (SBT). The known outbreaks and small clusters could be detected and clear discrimination of ST1 non-related isolates was obtained. In addition, the 50 loci cgMLST allowed to classify the isolates into subtypes because almost all the 50 genes could be called in all the analysed isolates, which was not achieved by the other tools. This is a big advantage in terms of standardisation and comparison between laboratories for future epidemiological investigations. WGS allowed to analyse a large volume of samples and generated more accurate conclusions for outbreak investigations compared to other typing methods due to its higher discriminatory power and throughput.
Keywords: Legionella pneumophila; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical statement: Epidemiological data were collected anonymously in the frame of Decision No 2119/98/EC of the European Parliament and of the Council, concerning the epidemiological surveillance and control of communicable diseases in the Community, as completed by Decision No 1082/2013/EU. Competing Interests: The authors declare no competing interests.
Figures

References
-
- Fraser DW (1980) Legionellosis: evidence of airborne transmission. Ann N Y Acad Sci 353:61–66. 10.1111/j.1749-6632.1980.tb18906.x - PubMed
-
- Fraser DW et al (1977) Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med 297(22):1189–1197. 10.1056/NEJM197712012972201 - PubMed
-
- Glick TH, Gregg MB, Berman B, Mallison G, Rhodes WW Jr, Kassanoff I (1978) Pontiac fever. An epidemic of unknown etiology in a health department: I. Clinical and epidemiologic aspects. Am J Epidemiol 107(2):149–160. 10.1093/oxfordjournals.aje.a112517 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

