Single-cell RNA sequencing-derived signatures define response patterns to atezolizumab + bevacizumab in advanced hepatocellular carcinoma
- PMID: 39709141
- PMCID: PMC12086051
- DOI: 10.1016/j.jhep.2024.12.016
Single-cell RNA sequencing-derived signatures define response patterns to atezolizumab + bevacizumab in advanced hepatocellular carcinoma
Abstract
Background & aims: The combination of atezolizumab and bevacizumab (atezo+bev) is the current standard of care for advanced hepatocellular carcinoma (HCC), providing a median overall survival (OS) of 19.2 months. Here, we aim to uncover the underlying cellular processes driving clinical benefit vs. resistance to atezo+bev.
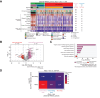
Methods: We harnessed the power of single-cell RNA sequencing in advanced HCC to derive gene expression signatures recapitulating 21 cell phenotypes. These signatures were applied to 422 RNA-sequencing samples of patients with advanced HCC treated with atezo+bev (n = 317) vs. atezolizumab (n = 47) or sorafenib (n = 58) as comparators.
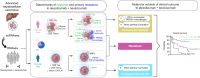
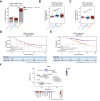
Results: We unveiled two distinct patterns of response to atezo+bev. First, an immune-mediated response characterised by the combined presence of CD8+ T effector cells and pro-inflammatory CXCL10+ macrophages, representing an immune-rich microenvironment. Second, a non-immune, angiogenesis-related response distinguishable by a reduced expression of the VEGF co-receptor neuropilin-1 (NRP1), a biomarker that specifically predicts improved OS upon atezo+bev vs. sorafenib (p = 0.039). Primary resistance was associated with an enrichment of immunosuppressive myeloid populations, namely CD14+ monocytes and TREM2+ macrophages, and Notch pathway activation. Based on these mechanistic insights we define "Immune-competent" and "Angiogenesis-driven" molecular subgroups, each associated with a significantly longer OS with atezo+bev vs. sorafenib (p of interaction = 0.027), and a "Resistant" subset.
Conclusion: Our study unveils two distinct molecular subsets of clinical benefit to atezolizumab plus bevacizumab in advanced HCC ("Immune-competent" and "Angiogenesis-driven") as well as the main traits of primary resistance to this therapy, thus providing a molecular framework to stratify patients based on clinical outcome and guiding potential strategies to overcome resistance.
Impact and implications: Atezolizumab + bevacizumab (atezo+bev) is standard of care in advanced hepatocellular carcinoma (HCC), yet molecular determinants of clinical benefit to the combination remain unclear. This study harnesses the power of single-cell RNA sequencing, deriving gene expression signatures representing 21 cell subtypes in the advanced HCC microenvironment. By applying these signatures to RNA-sequencing samples, we reveal two distinct response patterns to atezo+bev and define molecular subgroups of patients ("Immune-competent" and "Angiogenesis-driven" vs. "Resistant") with differential clinical outcomes upon treatment with atezo+bev, pointing towards the role of immunosuppressive myeloid cell types and Notch pathway activation in primary resistance to atezo+bev. These results may help refine treatment strategies and improve outcomes for patients with advanced HCC, while also guiding future research aimed at overcoming resistance mechanisms.
Keywords: Advanced Hepatocellular Carcinoma; Atezolizumab and bevacizumab; Biomarkers of Response; Primary Resistance; Single-Cell RNA-Sequencing.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest AD received educational support for congress attendance and consultancy fees from Roche, and speaker fees from Roche, AstraZeneca, Eisai, and Chugai. FFo has received honoraria for lectures from AstraZeneca, Lilly, MSD, Pfizer and Roche. He has served as advisory board member to AstraZeneca, BMS, Eisai and Roche and has received travel support from Merck KGaA and Servier. RM has received consulting and lecture fees from Servier, Roche and Bristol Myers Squibb and travel and education funding from MSD, Eli Lilly, Bayer, Roche, Astrazeneca. SG reports other research funding from Boehringer Ingelheim, Bristol-Myers Squibb, Celgene, Genentech, Regeneron, and Takeda not related to this study. SG is a named co-inventor on an issued patent for MICSSS, a multiplex immunohistochemistry to characterise tumours and treatment responses. The technology is filed through Icahn School of Medicine at Mount Sinai (ISMMS) and is currently unlicensed. AV has received consulting fees from FirstWorld, Natera, Pioneering Medicine and Genentech; advisory board fees from BMS, Roche, Astra Zeneca, Eisai, and NGM Pharmaceuticals; and research support from Eisai. He has stock options from Espervita. JML reports research support from Eisai Inc and Bayer Pharmaceuticals, consultancy/sponsored lectures from Eisai Inc., Merck, Roche, Genentech, AstraZeneca, Bayer Pharmaceuticals, Abbvie, Sanofi, Moderna, Glycotest and Exelixis, and Data Safety Monitoring Board for Industry or Commercial Enterprise from Bristol Myers Squibb. The remaining authors have no conflicts of interest to declare. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures





References
-
- Sung, Ferlay, Siegel, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Llovet, Ricci, Mazzaferro, et al. Sorafenib in advanced hepatocellular carcinoma. NEJM. 2008;359:378–390. - PubMed
-
- Finn, Qin, Ikeda, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. NEJM. 2020;382:1894–1905. - PubMed
-
- Cheng, Qin, Ikeda, et al. Updated efficacy and safety data from IMbrave150: atezolizumab plus bevacizumab vs. sorafenib for unresectable hepatocellular carcinoma. J Hepatol. 2022;76:862–873. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

