Transcriptional profile of the rat cardiovascular system at single-cell resolution
- PMID: 39709602
- PMCID: PMC11781962
- DOI: 10.1016/j.celrep.2024.115091
Transcriptional profile of the rat cardiovascular system at single-cell resolution
Abstract
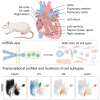
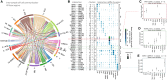
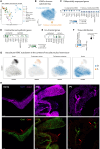
We sought to characterize cellular composition across the cardiovascular system of the healthy Wistar rat, an important model in preclinical cardiovascular research. We performed single-nucleus RNA sequencing (snRNA-seq) in 78 samples in 10 distinct regions, including the four chambers of the heart, ventricular septum, sinoatrial node, atrioventricular node, aorta, pulmonary artery, and pulmonary veins, which produced 505,835 nuclei. We identified 26 distinct cell types and additional subtypes, with different cellular composition across cardiac regions and tissue-specific transcription for each cell type. Several cell subtypes were region specific, including a subtype of vascular smooth muscle cells enriched in the large vasculature. We observed tissue-enriched cellular communication networks, including heightened Nppa-Npr1/2/3 signaling in the sinoatrial node. The existence of tissue-restricted cell types suggests regional regulation of cardiovascular physiology. Our detailed transcriptional characterization of each cell type offers the potential to identify novel therapeutic targets and improve preclinical models of cardiovascular disease.
Keywords: CP: Developmental biology; atrioventricular node; cardiovascular disease; cardiovascular system; cell-cell communication; genetics; heart; pulmonary vein; rat; single-nuclei RNA sequencing; single-nucleus RNA sequencing; sinoatrial node.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.K. is an employee of Bayer US LLC (a subsidiary of Bayer AG) and may own stock in Bayer AG. H.M. was an employee of the Broad Institute at the time of project completion and is now an employee of STEMCELL Technologies. A.-D.A., I.P., and C.M.S. were employees of Bayer US LLC (a subsidiary of Bayer AG) at the time of project completion. I.P. is now an employee at BioMarin Pharmaceuticals, Inc. A.-D.A. and C.M.S. are now full-time employees of Absci Corp. A.A. was an employee of the Broad Institute at the time of project completion and is now an employee of Bayer US LLC. G.G.-C. is a scientific cofounder of Riparian Pharmaceuticals. P.T.E. receives sponsored research support from Bayer AG, Bristol Myers Squibb, Pfizer. and Novo Nordisk; he has also served on advisory boards and/or consulted for Bayer AG.
Figures





Update of
-
Transcriptional profile of the rat cardiovascular system at single cell resolution.bioRxiv [Preprint]. 2023 Nov 16:2023.11.14.567085. doi: 10.1101/2023.11.14.567085. bioRxiv. 2023. Update in: Cell Rep. 2025 Jan 28;44(1):115091. doi: 10.1016/j.celrep.2024.115091. PMID: 38014050 Free PMC article. Updated. Preprint.
References
-
- Plass M., Solana J., Wolf F.A., Ayoub S., Misios A., Glažar P., Obermayer B., Theis F.J., Kocks C., Rajewsky N. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science. 2018;360 - PubMed
-
- Chaffin M., Papangeli I., Simonson B., Akkad A.D., Hill M.C., Arduini A., Fleming S.J., Melanson M., Hayat S., Kost-Alimova M., et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy. Nature. 2022;608:174–180. - PubMed
-
- Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

