Model organisms for investigating the functional involvement of NRF2 in non-communicable diseases
- PMID: 39709790
- PMCID: PMC11733061
- DOI: 10.1016/j.redox.2024.103464
Model organisms for investigating the functional involvement of NRF2 in non-communicable diseases
Erratum in
-
Corrigendum to "Model organisms for investigating the functional involvement of NRF2 in non-communicable diseases" [Redox Biol. 79 (2025) 103464].Redox Biol. 2025 Mar;80:103496. doi: 10.1016/j.redox.2025.103496. Epub 2025 Jan 19. Redox Biol. 2025. PMID: 39833011 Free PMC article. No abstract available.
Abstract
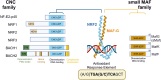
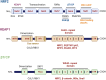
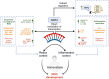
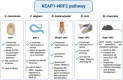
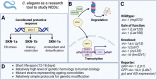
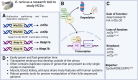
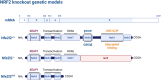
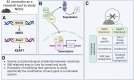
Non-communicable chronic diseases (NCDs) are most commonly characterized by age-related loss of homeostasis and/or by cumulative exposures to environmental factors, which lead to low-grade sustained generation of reactive oxygen species (ROS), chronic inflammation and metabolic imbalance. Nuclear factor erythroid 2-like 2 (NRF2) is a basic leucine-zipper transcription factor that regulates the cellular redox homeostasis. NRF2 controls the expression of more than 250 human genes that share in their regulatory regions a cis-acting enhancer termed the antioxidant response element (ARE). The products of these genes participate in numerous functions including biotransformation and redox homeostasis, lipid and iron metabolism, inflammation, proteostasis, as well as mitochondrial dynamics and energetics. Thus, it is possible that a single pharmacological NRF2 modulator might mitigate the effect of the main hallmarks of NCDs, including oxidative, proteostatic, inflammatory and/or metabolic stress. Research on model organisms has provided tremendous knowledge of the molecular mechanisms by which NRF2 affects NCDs pathogenesis. This review is a comprehensive summary of the most commonly used model organisms of NCDs in which NRF2 has been genetically or pharmacologically modulated, paving the way for drug development to combat NCDs. We discuss the validity and use of these models and identify future challenges.
Keywords: Inflammation; Model organisms; NRF2; Non-communicable chronic diseases; Oxidative stress.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Unwin N., Alberti K.G. Chronic non-communicable diseases. Ann. Trop. Med. Parasitol. 2006;100(5–6):455–464. - PubMed
-
- Tebay L.E., Robertson H., Durant S.T., Vitale S.R., Penning T.M., Dinkova-Kostova A.T., et al. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015;88(Pt B):108–146. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

