Chronic sleep deprivation disturbs energy balance modulated by suprachiasmatic nucleus efferents in mice
- PMID: 39710657
- PMCID: PMC11665105
- DOI: 10.1186/s12915-024-02097-4
Chronic sleep deprivation disturbs energy balance modulated by suprachiasmatic nucleus efferents in mice
Abstract
Background: Epidemiologic researches show that short sleep duration may affect feeding behaviors resulting in higher energy intake and increased risk of obesity, but the further mechanisms that can interpret the causality remain unclear. The circadian rhythm is fine-tuned by the suprachiasmatic nucleus (SCN) as the master clock, which is essential for driving rhythms in food intake and energy metabolism through neuronal projections to the arcuate nucleus (ARC) and paraventricular nucleus (PVN).
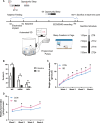
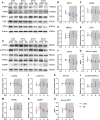
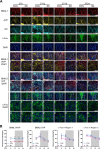
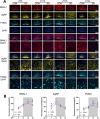
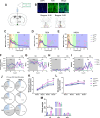
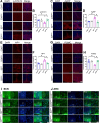
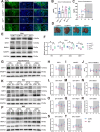
Results: We showed that chronic SD-induced aberrant expressions of AgRP/NPY and POMC attributed to compromised JAK/STAT3 signals and reduced energy expenditure in the mice, which can be rescued with AAV-genetic overexpression of BMAL1 into SCN. The potential mechanism may be related to the disruptions of SCN efferent mediated by BMAL1.
Conclusions: Chronic SD impairs energy balance through directly dampening BMAL1 expression, probably in the transcription level, in the SCN, which in turn affects the neuron projections to ARC and PVN. Remarkably, we provide evidence that may explain the causal mechanisms associated with sleep curtailment and obesity in adolescents.
Keywords: BMAL1; Chronic sleep deprivation; Energy expenditure; Food intake; SCN.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Ethics Committee of China Medical University (KT2022384), following the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Consent for publication: All authors consent for this manuscript to be published. Competing interests: The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

