Safflower CtFT genes orchestrating flowering time and flavonoid biosynthesis
- PMID: 39710673
- PMCID: PMC11665109
- DOI: 10.1186/s12870-024-05943-3
Safflower CtFT genes orchestrating flowering time and flavonoid biosynthesis
Abstract
Background: Safflower thrives in dry environments but faces difficulties with flowering in wet and rainy summers. Flavonoids play a role in flower development and can potentially alleviate these challenges. Furthermore, the FLOWERING LOCUS T (FT) family of phosphatidylethanolamine-binding protein (PEBP) genes play a crucial role in the photoperiodic flowering pathway. However, their direct impact on flowering and flavonoid biosynthesis under different light duration is elusive.
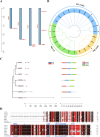
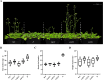
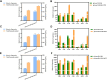
Results: Utilizing the genome sequencing of Safflower (Jihong NO.1), the current study identifies three specific genes (CtFT1, CtFT2, and CtFT3) that exhibit upregulation in response to long-day conditions. The overexpression of CtFT2, displayed an early, whereas CtFT1 and CtFT3 late flowering phenotype in Arabidopsis thaliana. Interestingly, the transient overexpression of CtFT1 in safflower leaves caused early flowering, while overexpressing CtFT2 and CtFT3 led to late flowering. Additionally, overexpressing CtFT3 in Arabidopsis and CtFT1, CtFT2, and CtFT3 in safflower leaves, significantly increased flavonoid synthesis.
Conclusions: These findings showed that overexpression of CtFT genes could affect the flowering time and significantly increase the flavonoid content of safflower. The function of CtFT gene is different in safflower and Arabidopsis. This study provides valuable insights into the role of CtFT genes in flower formation and flavonoid synthesis in safflower, which may help in improving safflower breeding quality and its adaptability to diverse environmental conditions.
Keywords: CtFTs; Flavonoid biosynthesis; Flower formation; Safflower.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. None of the species used in this study are endangered or protected, all plants were grown in greenhouses, and all experiments on these plants comply with all relevant guidelines and regulations. All plant materials were provided by Zhengzhou University. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Not applicable.
Figures



References
-
- Erbaş S, Mutlucan M. Investigation of flower yield and quality in different color safflower genotypes. Agronomy. 2023;13(4):956. - DOI
-
- Wang CC, Choy CS, Liu YH, Cheah KP, Li JS, Wang JTJ, Yu WY, Lin CW, Cheng HW, Hu CM. Protective effect of dried safflower petal aqueous extract and its main constituent, carthamus yellow, against lipopolysaccharide-induced inflammation in RAW264. 7 macrophages. J Sci Food Agric. 2011;91(2):218–25. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- . 20220204058YY, 20220505019ZP/Science and Technology Development Project of Jilin Province
- . 20220204058YY, 20220505019ZP/Science and Technology Development Project of Jilin Province
- 31771868/National Natural Science Foundation of China
- JJKH20230400KJ/Science and Technology Research Program of Education Department of Jilin province
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

