Human disease-causing missense genetic variants are enriched in the evolutionarily ancient domains of the cytosolic aminoacyl-tRNA synthetase proteins
- PMID: 39710895
- PMCID: PMC11664165
- DOI: 10.1002/iub.2932
Human disease-causing missense genetic variants are enriched in the evolutionarily ancient domains of the cytosolic aminoacyl-tRNA synthetase proteins
Abstract
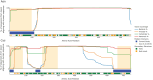
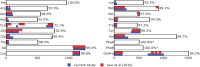
All life depends on accurate and efficient protein synthesis. The aminoacyl-tRNA synthetases (aaRSs) are a family of proteins that play an essential role in protein translation, as they catalyze the esterification reaction that charges a transfer RNA (tRNA) with its cognate amino acid. However, new domains added to the aaRSs over the course of evolution in eukaryotes confer novel functions unrelated to protein translation. To date, damaging variants that affect aaRS-encoding genes have been linked to over 50 human diseases. In this study, we leverage the evolutionary history of the aaRS proteins to better understand the distribution of disease-causing missense variants in human cytosolic aaRSs. We hypothesized that disease-causing missense variants in human aaRSs were more likely to be located in the ancient domains of the aaRS, essential for the aminoacylation reaction, rather than in the evolutionarily more recent domains found in eukaryotes. We determined the locations of the modern and ancient domains in each aaRS protein found in humans. We then statistically assessed the positional conservation across each domain and examined the distribution of pathogenic and benign/unknown missense human genetic variants across these domains. We establish that pathogenic missense variants in the human aaRS proteins are enriched in the evolutionarily ancient domains while benign/unknown missense variants are enriched in the modern domains. In addition to defining the evolutionary history of human aaRS proteins through domain identification, we anticipate that this work will improve the ability to diagnose patients affected by damaging genetic variants in the aaRS protein family.
Keywords: aminoacyl‐tRNA synthetases; conservation; evolution; genetic diseases; human health; protein synthesis; rare disease.
© 2024 The Author(s). IUBMB Life published by Wiley Periodicals LLC on behalf of International Union of Biochemistry and Molecular Biology.
Figures
Similar articles
-
Coexisting bacterial aminoacyl-tRNA synthetase paralogs exhibit distinct phylogenetic backgrounds and functional compatibility with Escherichia coli.IUBMB Life. 2024 Dec;76(12):1139-1153. doi: 10.1002/iub.2920. Epub 2024 Oct 17. IUBMB Life. 2024. PMID: 39417753
-
Photosynthetic demands on translational machinery drive retention of redundant tRNA metabolism in plant organelles.Proc Natl Acad Sci U S A. 2024 Dec 24;121(52):e2421485121. doi: 10.1073/pnas.2421485121. Epub 2024 Dec 18. Proc Natl Acad Sci U S A. 2024. PMID: 39693336 Free PMC article.
-
The IUBMB Focused Meeting on Aminoacyl-tRNA Synthetases 2023.IUBMB Life. 2025 Jul;77(7):e70041. doi: 10.1002/iub.70041. IUBMB Life. 2025. PMID: 40719009
-
Direct and quantitative analysis of tRNA acylation using intact tRNA liquid chromatography-mass spectrometry.Nat Protoc. 2025 May;20(5):1246-1274. doi: 10.1038/s41596-024-01086-9. Epub 2025 Jan 6. Nat Protoc. 2025. PMID: 39762443 Review.
-
The Lived Experience of Autistic Adults in Employment: A Systematic Search and Synthesis.Autism Adulthood. 2024 Dec 2;6(4):495-509. doi: 10.1089/aut.2022.0114. eCollection 2024 Dec. Autism Adulthood. 2024. PMID: 40018061 Review.
References
-
- Ibba M, Söll D. Aminoacyl‐tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. - PubMed
-
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier‐Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

