Tumour purity assessment with deep learning in colorectal cancer and impact on molecular analysis
- PMID: 39710952
- PMCID: PMC11717495
- DOI: 10.1002/path.6376
Tumour purity assessment with deep learning in colorectal cancer and impact on molecular analysis
Abstract
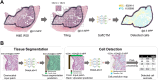
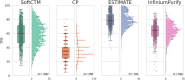
Tumour content plays a pivotal role in directing the bioinformatic analysis of molecular profiles such as copy number variation (CNV). In clinical application, tumour purity estimation (TPE) is achieved either through visual pathological review [conventional pathology (CP)] or the deconvolution of molecular data. While CP provides a direct measurement, it demonstrates modest reproducibility and lacks standardisation. Conversely, deconvolution methods offer an indirect assessment with uncertain accuracy, underscoring the necessity for innovative approaches. SoftCTM is an open-source, multiorgan deep-learning (DL) model for the detection of tumour and non-tumour cells in H&E-stained slides, developed within the Overlapped Cell on Tissue Dataset for Histopathology (OCELOT) Challenge 2023. Here, using three large multicentre colorectal cancer (CRC) cohorts (N = 1,097 patients) with digital pathology and multi-omic data, we compare the utility and accuracy of TPE with SoftCTM versus CP and bioinformatic deconvolution methods (RNA expression, DNA methylation) for downstream molecular analysis, including CNV profiling. SoftCTM showed technical repeatability when applied twice on the same slide (r = 1.0) and excellent correlations in paired H&E slides (r > 0.9). TPEs profiled by SoftCTM correlated highly with RNA expression (r = 0.59) and DNA methylation (r = 0.40), while TPEs by CP showed a lower correlation with RNA expression (r = 0.41) and DNA methylation (r = 0.29). We show that CP and deconvolution methods respectively underestimate and overestimate tumour content compared to SoftCTM, resulting in 6-13% differing CNV calls. In summary, TPE with SoftCTM enables reproducibility, automation, and standardisation at single-cell resolution. SoftCTM estimates (M = 58.9%, SD ±16.3%) reconcile the overestimation by molecular data extrapolation (RNA expression: M = 79.2%, SD ±10.5, DNA methylation: M = 62.7%, SD ±11.8%) and underestimation by CP (M = 35.9%, SD ±13.1%), providing a more reliable middle ground. A fully integrated computational pathology solution could therefore be used to improve downstream molecular analyses for research and clinics. © 2024 The Author(s). The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: artificial intelligence; colorectal cancer; diagnostic molecular pathology; pathology; personalised medicine.
© 2024 The Author(s). The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

