Influence of HLA-G 3' Untranslated Region Haplotypes and SNP +3422 Gene Variants as Host Genetic Factors on the Outcomes of SARS-CoV-2 Infection During Acute and Post-Acute Phases in a German Cohort
- PMID: 39711218
- PMCID: PMC11664307
- DOI: 10.1111/tan.15799
Influence of HLA-G 3' Untranslated Region Haplotypes and SNP +3422 Gene Variants as Host Genetic Factors on the Outcomes of SARS-CoV-2 Infection During Acute and Post-Acute Phases in a German Cohort
Abstract
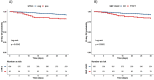
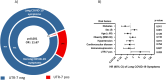
HLA-G, an important immune-checkpoint (IC) molecule that exerts inhibitory signalling on immune effector cells, has been suggested to represent a key player in regulating the immune response to Severe Acute Respiratory Syndrome Coronavirus Type 2 (SARS-CoV-2). Since specific single-nucleotide polymorphisms (SNP) in the HLA-G 3'untranslated region (UTR), which arrange as haplotypes, are crucial for the regulation of HLA-G expression, we analysed the contribution of these genetic variants as host factors in SARS-CoV-2 infection during acute and post-acute phases. HLA-G gene polymorphisms in the 3'UTR were investigated by sequencing in an unvaccinated Coronavirus Disease 2019 (COVID-19) cohort during acute SARS-CoV-2 infection (N = 505) and in the post-acute phase (N = 253). The HLA-G 3'UTR haplotype known as UTR-3 (p = 0.002) and the variant rs17875408 (also known as +3422) T variant (p = 0.004) are independent prognostic risk factors for fatal COVID-19. The +3422T variant (p = 0.006) predicted also the early loss of neutralising SARS-CoV-2 antibodies. In addition, the HLA-G 3'UTR haplotype UTR-7 (p = 0.023) emerged as an independent prognostic factor for increased susceptibility to Long-COVID symptoms after SARS-CoV-2 infection. Our study highlights that due to the variability of the 3'UTR genetic background, HLA-G has the potential to contribute to the progression of SARS-CoV-2 infection, extending to the development of Long-COVID symptoms, despite the likely alterations in the microenvironment and associated HLA-G-specific regulatory elements over the course of the disease. By spotlighting HLA-G, the importance of the genetic background of IC and their pivotal role in modulating immune responses during and after COVID-19 are emphasised.
Keywords: COVID‐19; HLA‐G; HLA‐G 3′UTR; SARS‐CoV‐2; long‐COVID.
© 2024 The Author(s). HLA: Immune Response Genetics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Initiative C.‐H. G., “The COVID‐19 Host Genetics Initiative, a Global Initiative to Elucidate the Role of Host Genetic Factors in Susceptibility and Severity of the SARS‐CoV‐2 Virus Pandemic,” European Journal of Human Genetics 28, no. 6 (2020): 715–718, 10.1038/s41431-020-0636-6. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

