Identification of ALDH7A1 as a DNA-methylation-driven gene in lung squamous cell carcinoma
- PMID: 39711312
- PMCID: PMC11703541
- DOI: 10.1080/07853890.2024.2442529
Identification of ALDH7A1 as a DNA-methylation-driven gene in lung squamous cell carcinoma
Abstract
Background: Deoxyribose nucleic acid (DNA) methylation is an important epigenetic modification that plays an important role in the occurrence and development of tumors. Identifying key methylation-driven genes that affect the prognosis of lung squamous cell carcinoma (LUSC) can provide direction for targeted therapy research.
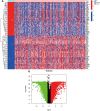
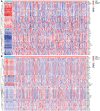
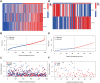
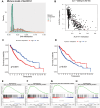
Methods and results: Methylation and RNA-seq data were downloaded from The Cancer Genome Atlas (TCGA). The MethylMix package was used to integrate and analyze the methylation and gene expression data from TCGA, and the LUSC dataset (GSE37745) was downloaded from GEO for validation. Forty-five DNA-methylation-driven genes (MDGs) were obtained, and 3 genes (TRIM61, SMIM22, and ALDH7A1) were significantly associated with survival by using univariate and multivariate Cox regression. A risk model was constructed. KM analysis showed that patients with high-risk scores had poor survival. A nomination plot for prognosis prediction of LUSC patients was constructed, which showed a good predictive efficiency for tumor prognosis. The high expression of ALDH7A1 was an independent risk factor for poor prognosis in LUSC. The expression of ALDH7A1 in LUSC was negatively correlated with its methylation status (COR = -0.655). GSEA analysis showed that high expression of ALDH7A1 could activate multiple signaling pathways (JAK-STAT signaling pathway and mTOR signaling pathway). In vitro cell experiments confirmed that in LUSC, silencing ALDH7A1 could inhibit tumor progression, while overexpression of ALDH7A1 could promote tumor progression.
Conclusion: Our results indicated that ALDH7A1, a newly discovered MDG in LUSC, could act as an independent prognostic factor for OS in LUSC, with the potential to become a potential target for LUSC diagnosis and treatment. High expression of ALDH7A1 in LUSC could promote the occurrence and development of tumors. Signaling pathways, such as JAK-STAT and mTOR signaling pathways, might regulate the high expression of ALDH7A1.
Keywords: ALDH7A; DNA methylation; Lung squamous cell carcinoma; methylation driver gene; survival analysis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




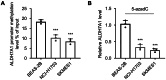
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
