This is a preprint.
CYP1B1-AS1 regulates CYP1B1 to promote Coxiella burnetii pathogenesis by inhibiting ROS and host cell death
- PMID: 39711545
- PMCID: PMC11661375
- DOI: 10.21203/rs.3.rs-5390645/v1
CYP1B1-AS1 regulates CYP1B1 to promote Coxiella burnetii pathogenesis by inhibiting ROS and host cell death
Update in
-
CYP1B1-AS1 regulates CYP1B1 to promote Coxiella burnetii pathogenesis by inhibiting ROS and host cell death.Nat Commun. 2025 Aug 13;16(1):7493. doi: 10.1038/s41467-025-62762-2. Nat Commun. 2025. PMID: 40796858 Free PMC article.
Abstract
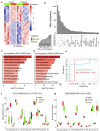
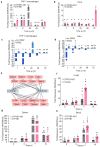
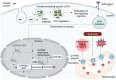
Coxiella burnetii (Cb), the causative agent of Q fever, replicates within host macrophages by modulating immune responses through poorly understood mechanisms. Long non-coding RNAs (lncRNAs) are emerging as critical regulators of inflammation, yet their role in Cb pathogenesis remains largely unexplored. Here, we employed a global transcriptomic approach to identify lncRNAs specific to Cb infection in THP-1 derived macrophages, compared to 15 other microbial infections. CYP1B1-AS1 was uniquely regulated in a spatio-temporal manner during Cb infection. Promoter assays revealed that CYP1B1-AS1 is transcribed by AHR from a bidirectional promoter, enhancing CYP1B1 expression in cis. Inhibition of CYP1B1-AS1 and CYP1B1 increased reactive oxygen species (ROS), mitochondrial membrane depolarization, and apoptosis, suggesting their role in dampening host cell death. Additionally, immunoprecipitation followed by mass spectrometry identified the mitochondrially localized Cb effector CBU_0937 as an interactor of the CYP1B1 enzyme. These events facilitate Cb intracellular survival. Our findings identify CYP1B1-AS1 as a potential molecular target for combating Cb infection.
Keywords: CBU_0937; CYP1B1; CYP1B1-AS1; Coxiella burnetii; apoptosis; bacterial pathogenesis; inflammation; lncRNA marker; mitochondrial effector; reactive oxygen species (ROS).
Conflict of interest statement
Competing interests The authors declare no other competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

