Temporal dynamics in a red alga dominated geothermal feature in Yellowstone National Park
- PMID: 39711979
- PMCID: PMC11662350
- DOI: 10.1093/ismeco/ycae151
Temporal dynamics in a red alga dominated geothermal feature in Yellowstone National Park
Abstract
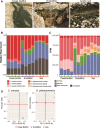
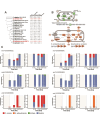
Alga-dominated geothermal spring communities in Yellowstone National Park (YNP), USA, have been the focus of many studies, however, relatively little is known about the composition and community interactions which underpin these ecosystems. Our goal was to determine, in three neighboring yet distinct environments in Lemonade Creek, YNP, how cells cope with abiotic stressors over the diurnal cycle. All three environments are colonized by two photosynthetic lineages, Cyanidioschyzon and Galdieria, both of which are extremophilic Cyanidiophyceae red algae. Cyanidioschyzon, a highly specialized obligate photoautotroph, dominated cell counts at all three Lemonade Creek environments. The cell cycle of Cyanidioschyzon in YNP matched that observed in synchronized cultures, suggesting that light availability plays a strong role in constraining growth of this alga in its natural habitat. Surprisingly, the mixotrophic and physiologically more flexible Galdieria, was a minor component of these algal populations. Arsenic detoxification at Lemonade Creek occurred via complementary gene expression by different eukaryotic and prokaryotic lineages, consistent with this function being shared by the microbial community, rather than individual lineages completing the entire pathway. These results demonstrate the highly structured nature of these extreme habitats, particularly regarding arsenic detoxification.
Keywords: Yellowstone National Park; community interactions; cyanidiophyceae; extremophiles; hot springs; microbiome; multi-omics.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
Authors declare that they have no competing interests.
Figures


References
-
- Castenholz RW, McDermott TR. The Cyanidiales: Ecology, biodiversity, and biogeography. In: Seckbach J., Chapman D.J. (eds.), Red Algae in the Genomic Age. Netherlands, Dordrecht: Springer, 2010, 357–71. 10.1007/978-90-481-3795-4_19 - DOI
-
- Reeb V, Bhattacharya D. The Thermo-acidophilic Cyanidiophyceae (Cyanidiales). In: Seckbach J., Chapman D.J. (eds.), Red Algae in the Genomic Age. Netherlands, Dordrecht: Springer, 2010, 409–26. 10.1007/978-90-481-3795-4_22 - DOI
LinkOut - more resources
Full Text Sources

