Unravelling turbot (Scophthalmus maximus) resistance to Aeromonas salmonicida: transcriptomic insights from two full-sibling families with divergent susceptibility
- PMID: 39712009
- PMCID: PMC11659141
- DOI: 10.3389/fimmu.2024.1522666
Unravelling turbot (Scophthalmus maximus) resistance to Aeromonas salmonicida: transcriptomic insights from two full-sibling families with divergent susceptibility
Abstract
Introduction: Furunculosis, caused by the gram-negative bacterium Aeromonas salmonicida subsp. salmonicida, remains a significant threat to turbot (Scophthalmus maximus) aquaculture. Identifying genetic backgrounds with enhanced disease resistance is critical for improving aquaculture health management, reducing antibiotic dependency, and mitigating economic losses.
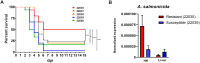
Methods: In this study, five full-sibling turbot families were challenged with A. salmonicida, which revealed one family with significantly greater resistance. Transcriptomic analyses (RNA-Seq) were performed on resistant and susceptible families, examining both naïve and 24-h postinfection (hpi) samples from head kidney and liver tissues.
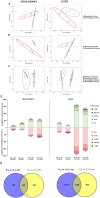
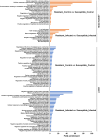
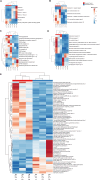
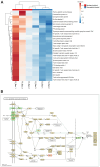
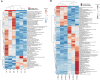
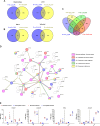
Results: In the absence of infection, differentially expressed genes (DEGs) were identified predominantly in the liver. Following infection, a marked increase in DEGs was observed in the head kidney, with many genes linked to immune functions. Interestingly, the resistant family displayed a more controlled inflammatory response and upregulation of genes related to antigen presentation and T-cell activity in the head kidney at early infection stages, which may have contributed to its increased survival rate. In the liver, transcriptomic differences between the families were associated mainly with cytoskeletal organization, cell cycle regulation, and metabolic processes, including insulin signalling and lipid metabolism, regardless of infection status. Additionally, many DEGs overlapped with previously identified quantitative trait loci (QTLs) associated with resistance to A. salmonicida, providing further insights into the genetic basis of disease resistance.
Discussion: This study represents the first RNA-Seq analysis comparing resistant and susceptible turbot families and contributes valuable knowledge for the development of selective breeding programs targeting disease resistance in turbot and other aquaculture species susceptible to A. salmonicida.
Keywords: Aeromonas salmonicida; disease resistance; furunculosis; transcriptome sequencing; turbot families.
Copyright © 2024 Pereiro, Tur, García, Figueras and Novoa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




Similar articles
-
Comparison of Aeromonas salmonicida resistant and susceptible salmon families: a high immune response is beneficial for the survival against Aeromonas salmonicida challenge.Fish Shellfish Immunol. 2011 Jul;31(1):1-9. doi: 10.1016/j.fsi.2010.12.019. Epub 2011 Jan 11. Fish Shellfish Immunol. 2011. PMID: 21232605
-
QTL detection for Aeromonas salmonicida resistance related traits in turbot (Scophthalmus maximus).BMC Genomics. 2011 Nov 2;12:541. doi: 10.1186/1471-2164-12-541. BMC Genomics. 2011. PMID: 22047500 Free PMC article.
-
Gene expression profiles of the spleen, liver, and head kidney in turbot (Scophthalmus maximus) along the infection process with Aeromonas salmonicida using an immune-enriched oligo-microarray.Mar Biotechnol (NY). 2011 Dec;13(6):1099-114. doi: 10.1007/s10126-011-9374-7. Epub 2011 Apr 19. Mar Biotechnol (NY). 2011. PMID: 21503602
-
Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis.Vet Microbiol. 2014 Feb 21;169(1-2):1-7. doi: 10.1016/j.vetmic.2013.06.025. Epub 2013 Jul 9. Vet Microbiol. 2014. PMID: 23890675 Review.
-
Aeromonas salmonicida: updates on an old acquaintance.Dis Aquat Organ. 2016 Jun 15;120(1):49-68. doi: 10.3354/dao03006. Dis Aquat Organ. 2016. PMID: 27304870 Review.
Cited by
-
Influence of Viral Re-Infection on Head Kidney Transcriptome of Nervous Necrosis Virus-Resistant and -Susceptible European Sea Bass (Dicentrarchus labrax, L.).Viruses. 2025 Feb 7;17(2):230. doi: 10.3390/v17020230. Viruses. 2025. PMID: 40006985 Free PMC article.
-
Integrated transcriptomic and microbiomic analyses reveal mechanisms of Decapod iridescent virus 1 resistance in Macrobrachium rosenbergii.Front Immunol. 2025 May 26;16:1611481. doi: 10.3389/fimmu.2025.1611481. eCollection 2025. Front Immunol. 2025. PMID: 40491929 Free PMC article.
References
-
- APROMAR . Aquaculture in Spain 2023(2023). Available online at: https://apromar.es/wp-content/uploads/2023/10/Aquaculture_in_Spain_2023_... (accessed November 27, 2024).
-
- FAO . The state of world fisheries and aquaculture 2022. In: Towards Blue Transformation. FAO, Rome: (2022). doi: 10.4060/cc0461en - DOI
-
- Nougayrede P, Sochon E, Vuiiaume A. Isolation of Aeromonas subspecies salmonicida in farmed turbot (Psetta maxima) in France. Bull Eur Assoc Fish Pathol. (1990) 10:139–40.
-
- Toranzo AE, Barja JL. First report of furunculosis in turbot reared in floating cages in Northwest Spain. Bull Eur Ass Fish Pathol. (1992) 12:147–9.
MeSH terms
LinkOut - more resources
Full Text Sources

