Exploring the heterogeneity of osteosarcoma cell characteristics and metabolic states and their association with clinical prognosis
- PMID: 39712023
- PMCID: PMC11659294
- DOI: 10.3389/fimmu.2024.1507476
Exploring the heterogeneity of osteosarcoma cell characteristics and metabolic states and their association with clinical prognosis
Abstract
Background: Osteosarcoma is a malignant tumor originating from mesenchymal bone tissue, characterized by high malignancy and poor prognosis. Despite progress in comprehensive treatment approaches, the five-year survival rate remains largely unchanged, highlighting the need to clarify its underlying mechanisms and discover new therapeutic targets.
Methods: This study utilized RNA sequencing data from multiple public databases, encompassing osteosarcoma samples and healthy controls, along with single-cell RNA sequencing data. Various methods were utilized, such as differential expression analysis of genes, analysis of metabolic pathways, and weighted gene co-expression network analysis (WGCNA), to pinpoint crucial genes. Using this list of genes, we developed and validated a prognostic model that incorporated risk signatures, and we evaluated the effectiveness of the model through survival analysis, immune cell infiltration examination, and drug sensitivity evaluation.
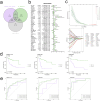
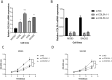
Results: We analyzed gene expression and metabolic pathways in nine samples using single-cell sequencing data. Initially, we performed quality control and clustering, identifying 21 statistically significant cell subpopulations. Metabolic analyses of these subpopulations revealed heterogeneous activation of metabolic pathways. Focusing on the osteoblastic cell subpopulation, we further subdivided it into six groups and examined their gene expression and differentiation capabilities. Differential expression and enrichment analyses indicated that tumor tissues were enriched in cytoskeletal and structural pathways. Through WGCNA, we identified core genes negatively correlated with four highly activated metabolic pathways. Using osteosarcoma patient data, we developed a risk signature model that demonstrated robust prognostic predictions across three independent cohorts. Ultimately, we performed a thorough examination of the model, which encompassed clinical and pathological characteristics, enrichment analysis, pathways associated with cancer markers, and scores of immune infiltration, highlighting notable and complex disparities between high-risk and low-risk populations.
Conclusion: This research clarifies the molecular mechanisms and metabolic features associated with osteosarcoma and how they relate to patient outcomes, offering novel perspectives and approaches for targeted therapy and prognostic assessment in osteosarcoma.
Keywords: comprehensive analysis; immune infiltration; metabolic pathways; osteosarcoma; prognostic analysis.
Copyright © 2024 Qin, Hu, Deng and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








Similar articles
-
Comprehensive investigation of tumor immune microenvironment and prognostic biomarkers in osteosarcoma through integrated bulk and single-cell transcriptomic analysis.Front Immunol. 2024 Jul 23;15:1424950. doi: 10.3389/fimmu.2024.1424950. eCollection 2024. Front Immunol. 2024. PMID: 39108264 Free PMC article.
-
Develop a Novel Signature to Predict the Survival and Affect the Immune Microenvironment of Osteosarcoma Patients: Anoikis-Related Genes.J Immunol Res. 2024 Mar 27;2024:6595252. doi: 10.1155/2024/6595252. eCollection 2024. J Immunol Res. 2024. PMID: 39431237 Free PMC article.
-
Glypican-3 regulated epithelial mesenchymal transformation-related genes in osteosarcoma: based on comprehensive tumor microenvironment profiling.Front Immunol. 2025 May 13;16:1566061. doi: 10.3389/fimmu.2025.1566061. eCollection 2025. Front Immunol. 2025. PMID: 40433364 Free PMC article.
-
Harnessing multi‑omics to revolutionize understanding and management of osteosarcoma: A pathway to precision medicine (Review).Int J Mol Med. 2025 Jun;55(6):92. doi: 10.3892/ijmm.2025.5533. Epub 2025 Apr 17. Int J Mol Med. 2025. PMID: 40242955 Free PMC article. Review.
-
Building a Therapeutic Bridge Between Dogs and Humans: A Review of Potential Cross-Species Osteosarcoma Biomarkers.Int J Mol Sci. 2025 May 28;26(11):5152. doi: 10.3390/ijms26115152. Int J Mol Sci. 2025. PMID: 40507962 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

