Mycobiome analysis of leaf, root, and soil of symptomatic oil palm trees (Elaeis guineensis Jacq.) affected by leaf spot disease
- PMID: 39712899
- PMCID: PMC11659247
- DOI: 10.3389/fmicb.2024.1422360
Mycobiome analysis of leaf, root, and soil of symptomatic oil palm trees (Elaeis guineensis Jacq.) affected by leaf spot disease
Abstract
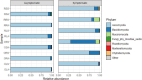
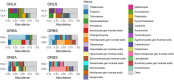
Recently, attention has been shifting toward the perspective of the existence of plants and microbes as a functioning ecological unit. However, studies highlighting the impacts of the microbial community on plant health are still limited. In this study, fungal community (mycobiome) of leaf, root, and soil of symptomatic leaf-spot diseased (SS) oil palm were compared against asymptomatic (AS) trees using ITS2 rRNA gene metabarcoding. A total of 3,435,417 high-quality sequences were obtained from 29 samples investigated. Out of the 14 phyla identified, Ascomycota and Basidiomycota were the most dominant accounting for 94.2 and 4.7% of the total counts in AS, and 75 and 21.2% in SS, respectively. Neopestalotiopsis is the most abundant genus for AS representing 8.0% of the identified amplicons compared to 2.0% in SS while Peniophora is the most abundant with 8.6% of the identified amplicons for SS compared to 0.1% in AS. The biomarker discovery algorithm LEfSe revealed different taxa signatures for the sample categories, particularly soil samples from asymptomatic trees, which were the most enriched. Network analysis revealed high modularity across all groups, except in root samples. Additionally, a large proportion of the identified keystone species consisted of rare taxa, suggesting potential role in ecosystem functions. Surprisingly both AS and SS leaf samples shared taxa previously associated with oil palm leaf spot disease. The significant abundance of Trichoderma asperellum in the asymptomatic root samples could be further explored as a potential biocontrol agent against oil palm disease.
Keywords: asymptomatic; fungal community; leaf spot disease; mycobiome; oil palm; symptomatic.
Copyright © 2024 Azeez, Esiegbuya, Lateef and Asiegbu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures






References
-
- Acevedo E., Galindo-Castañeda T., Prada F., Navia M., Romero H. (2014). Phosphate-solubilizing microorganisms associated with the rhizosphere of oil palm (Elaeis guineensis Jacq.) in Colombia. Appl. Soil Ecol. 80 26–33. 10.1016/j.apsoil.2014.03.011 - DOI
-
- Aderungboye F. (1977). Diseases of the oil palm. PANS 23 305–326. 10.1080/09670877709412457 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

