A ubiquinone precursor analogue does not clearly increase the growth rate of Caenorhabditis inopinata
- PMID: 39712935
- PMCID: PMC11659882
- DOI: 10.17912/micropub.biology.001235
A ubiquinone precursor analogue does not clearly increase the growth rate of Caenorhabditis inopinata
Abstract
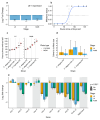
The evolution of developmental rates may drive morphological change. Caenorhabditis inopinata develops nearly twice as slowly as Caenorhabditis elegans . clk-1 encodes a hydroxylase required for synthesizing ubiquinone, and mutant clk-1 slow growth phenotypes can be rescued by supplying animals with a ubiquinone precursor analogue, 2,4-dihydroxybenzoate. RNA-seq data showing low clk-1 expression raised the possibility that C. inopinata grows slowly because of reduced ubiquinone biosynthesis. C. inopinata did not reveal a clear reduction in the age of maturation when reared on 2,4-dihydroxybenzoate. Further scrutiny of RNA-seq results revealed multiple ubiquinone metabolism genes have low expression in C. inopinata . Divergent clk-1 expression alone may not be a major driver of the evolution of slow development in this species.
Copyright: © 2024 by the authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest present.
Figures
References
-
- Alberch Pere, Gould Stephen Jay, Oster George F., Wake David B. Size and shape in ontogeny and phylogeny. Paleobiology. 1979;5(3):296–317. doi: 10.1017/s0094837300006588. - DOI
-
- Benjamini Yoav, Hochberg Yosef. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B: Statistical Methodology. 1995 Jan 1;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. - DOI
-
- Calder WA. 1984. Size, function, and life history . Cambridge (MA): Harvard University Press.
-
- Dirksen Philipp, Marsh Sarah Arnaud, Braker Ines, Heitland Nele, Wagner Sophia, Nakad Rania, Mader Sebastian, Petersen Carola, Kowallik Vienna, Rosenstiel Philip, Félix Marie-Anne, Schulenburg Hinrich. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biology. 2016 May 9;14(1) doi: 10.1186/s12915-016-0258-1. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
