Unveiling the Mechanism of Retinoic Acid Therapy for Cutaneous Warts: Insights from Multi-Omics Integration
- PMID: 39712939
- PMCID: PMC11663382
- DOI: 10.2147/CCID.S504391
Unveiling the Mechanism of Retinoic Acid Therapy for Cutaneous Warts: Insights from Multi-Omics Integration
Abstract
Background: Due to limited treatment options, cutaneous warts caused by human papillomavirus (HPV) remain a significant clinical challenge. Furthermore, the genetic susceptibility and molecular basis of viral warts are not yet fully understood.
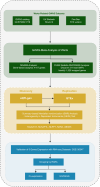
Methods: We utilized a multi-omics integration approach, encompassing genome-wide association study (GWAS) meta-analysis, summary data-based Mendelian randomization (SMR) analysis, and transcriptomic validation using the GSE136347 dataset. Differential gene expression (DEG) analysis was conducted to identify significant changes in gene expression between wart tissues and healthy skin.
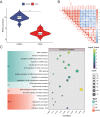
Results: Our analyses revealed five genetic susceptibility genes associated with cutaneous warts, with RARA showing significant differential expression in wart tissues. Co-expression analysis indicated that RARA may regulate apoptosis through interactions with BAX, a pro-apoptotic gene. Additionally, functional annotation via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses highlighted key biological processes and pathways involved in wart pathogenesis.
Conclusion: This study identifies RARA as a pivotal regulator in the molecular pathology of cutaneous warts and a promising therapeutic target. RA-based therapies could offer effective and less invasive alternatives for wart treatment. Future investigations should refine the molecular role of RARA to optimize clinical interventions.
Keywords: GWAS-meta; RARA; cutaneous warts; multi-omics; retinoic acid.
© 2024 Dong et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures

References
LinkOut - more resources
Full Text Sources
Medical
Research Materials

