Mutant huntingtin protein decreases with CAG repeat expansion: implications for therapeutics and bioassays
- PMID: 39713241
- PMCID: PMC11660907
- DOI: 10.1093/braincomms/fcae410
Mutant huntingtin protein decreases with CAG repeat expansion: implications for therapeutics and bioassays
Abstract
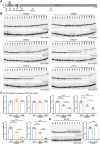
Huntington's disease is an inherited neurodegenerative disorder caused by a CAG repeat expansion that encodes a polyglutamine tract in the huntingtin (HTT) protein. The mutant CAG repeat is unstable and expands in specific brain cells and peripheral tissues throughout life. Genes involved in the DNA mismatch repair pathways, known to act on expansion, have been identified as genetic modifiers; therefore, it is the rate of somatic CAG repeat expansion that drives the age of onset and rate of disease progression. In the context of an expanded CAG repeat, the HTT pre-mRNA can be alternatively processed to generate the HTT1a transcript that encodes the aggregation prone and highly pathogenic HTT1a protein. This may be a mechanism through which somatic CAG repeat expansion exerts its pathogenic effects, as the longer the CAG repeat, the more HTT1a and HTT1a is produced. The allelic series of knock-in mouse models, HdhQ20, HdhQ50, HdhQ80, HdhQ111, CAG140 and zQ175 with polyglutamine expansions of 20, 50, 80, 111, 140 and ∼190, can be used to model the molecular and cellular consequences of CAG repeat expansion within a single neuron. By western blot of cortical lysates, we found that mutant HTT levels decreased with increasing CAG repeat length; mutant HTT was only 23 and 10% of wild-type levels in CAG140 and zQ175 cortices, respectively. To identify the optimal bioassays for detecting the full-length HTT and HTT1a isoforms, we interrogated the pairwise combinations of seven well-characterized antibodies on both the 'homogeneous time-resolved fluorescence' and 'Meso Scale Discovery' platforms. We tested 32 assays on each platform to detect 'full-length mutant HTT', HTT1a, 'total mutant HTT' (full-length HTT and HTT1a) and 'total full-length HTT' (mutant and wild type). None of these assays recapitulated the full-length mutant HTT levels as measured by western blot. We recommend using isoform- and species-specific assays that detect full-length mutant HTT, HTT1a or wild-type HTT as opposed to those that detect more than one isoform simultaneously. Our finding that as the CAG repeat expands, full-length mutant HTT levels decrease, while HTT1a and HTT1a levels increase has implications for therapeutic strategies. If mutant HTT levels in cells containing (CAG)200 are only 10% of wild-type, HTT-lowering strategies targeting full-length HTT at sequences 3' to Intron 1 HTT will predominantly lower wild-type HTT, as mutant HTT levels in these cells are already depleted. These data support a therapeutic strategy that lowers HTT1a and depletes levels of the HTT1a protein.
Keywords: HTT1a protein; Huntington’s disease; huntingtin bioassay; knock-in mouse models of Huntington’s disease; mutant huntingtin protein.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
J.R.G. and K.B. are employees of Rancho BioSciences. The authors report no other competing interests.
Figures







References
-
- Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat Rev Dis Primers. 2015;1:15005. - PubMed
-
- HDCRG . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–983. - PubMed
-
- Duyao M, Ambrose C, Myers R, et al. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet. 1993;4(4):387–392. - PubMed
-
- Telenius H, Kremer HP, Theilmann J, et al. Molecular analysis of juvenile Huntington disease: The major influence on (CAG)n repeat length is the sex of the affected parent. Hum Mol Genet. 1993;2(10):1535–1540. - PubMed
-
- Telenius H, Kremer B, Goldberg YP, et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm [published erratum appears in Nat Genet 1994 May;7(1):113]. Nat Genet. 1994;6(4):409–414. - PubMed
LinkOut - more resources
Full Text Sources
