This is a preprint.
Bicoid-nucleosome competition sets a concentration threshold for transcription constrained by genome replication
- PMID: 39713295
- PMCID: PMC11661180
- DOI: 10.1101/2024.12.10.627802
Bicoid-nucleosome competition sets a concentration threshold for transcription constrained by genome replication
Update in
-
Bicoid-nucleosome competition sets a concentration threshold for transcription constrained by genome replication.Cell Rep. 2025 Aug 26;44(8):116121. doi: 10.1016/j.celrep.2025.116121. Epub 2025 Aug 6. Cell Rep. 2025. PMID: 40779393 Free PMC article.
Abstract
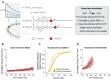
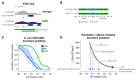
Transcription factors (TFs) regulate gene expression despite constraints from chromatin structure and the cell cycle. Here we examine the concentration-dependent regulation of hunchback by the Bicoid morphogen through a combination of quantitative imaging, mathematical modeling and epigenomics in Drosophila embryos. By live imaging of MS2 reporters, we find that, following mitosis, the timing of transcriptional activation driven by the hunchback P2 (hb P2) enhancer directly reflects Bicoid concentration. We build a stochastic model that can explain in vivo onset time distributions by accounting for both the competition between Bicoid and nucleosomes at hb P2 and a negative influence of DNA replication on transcriptional elongation. Experimental modulation of nucleosome stability alters onset time distributions and the posterior boundary of hunchback expression. We conclude that TF-nucleosome competition is the molecular mechanism whereby the Bicoid morphogen gradient specifies the posterior boundary of hunchback expression.
Figures





References
-
- Boijja A., Klein I. A., Sabari B. R., Dall’Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., & Young R. A. (2018). Transcription factors activate genes through the phase separation capacity of their activation domains. Cell, 175(7), 1842–1855.e16. 10.1016/j.cell.2018.10.042 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
