This is a preprint.
The CECR2 bromodomain displays distinct binding modes to select for acetylated histone proteins versus non-histone ligands
- PMID: 39713312
- PMCID: PMC11661176
- DOI: 10.1101/2024.12.09.627393
The CECR2 bromodomain displays distinct binding modes to select for acetylated histone proteins versus non-histone ligands
Abstract
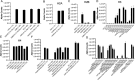
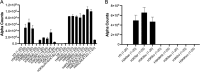
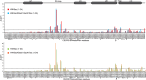
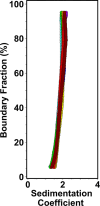
The cat eye syndrome chromosome region candidate 2 (CECR2) protein is an epigenetic regulator involved in chromatin remodeling and transcriptional control. The CECR2 bromodomain (CECR2-BRD) plays a pivotal role in directing the activity of CECR2 through its capacity to recognize and bind acetylated lysine residues on histone proteins. This study elucidates the binding specificity and structural mechanisms of CECR2-BRD interactions with both histone and non-histone ligands, employing techniques such as isothermal titration calorimetry (ITC), nuclear magnetic resonance (NMR) spectroscopy, and a high-throughput peptide assay. The CECR2-BRD selectively binds acetylated histone H3 and H4 ligands, exhibiting a preference for multi-acetylated over mono-acetylated targets. The highest affinity was observed for tetra-acetylated histone H4. Neighboring post-translational modifications, including methylation and phosphorylation, modulate acetyllysine recognition, with significant effects observed for histone H3 ligands. Additionally, this study explored the interaction of the CECR2-BRD with the acetylated RelA subunit of NF-κB, a pivotal transcription factor in inflammatory signaling. Dysregulated NF-κB signaling is implicated in numerous pathologies, including cancer progression, with acetylation of RelA at lysine 310 (K310ac) being critical for its transcriptional activity. Recent evidence linking the CECR2-BRD to RelA suggests it plays a role in inflammatory and metastatic pathways, underscoring the need to understand the molecular basis of this interaction. We found the CECR2-BRD binds to acetylated RelA with micromolar affinity, and uses a distinctive binding mode to recognize this non-histone ligand. These results provide new insight on the role of CECR2 in regulating NF-κB-mediated inflammatory pathways. Functional mutagenesis of critical residues, such as Asn514 and Asp464, highlight their roles in ligand specificity and binding dynamics. Notably, the CECR2-BRD remained monomeric in solution and exhibited differential conformational responses upon ligand binding, suggesting adaptive recognition mechanisms. Furthermore, the CECR2-BRD exclusively interacts with nucleosome substrates containing multi-acetylated histones, emphasizing its role in transcriptional activation within euchromatic regions. These findings position the CECR2-BRD as a key chromatin reader and a promising therapeutic target for modulating transcriptional and inflammatory processes, particularly through the development of selective bromodomain inhibitors.
Conflict of interest statement
COMPETING INTERESTS: EpiCypher (M.R.M. and L.K.) is a commercial developer of the dCypher peptide-binding platform used in this study.
Figures
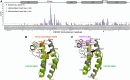


References
-
- Banting G. S.; Barak O.; Ames T. M.; Burnham A. C.; Kardel M. D.; Cooch N. S.; Davidson C. E.; Godbout R.; McDermid H. E.; Shiekhattar R. CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum Mol Genet 2005, 14 (4), 513–524. DOI: 10.1093/hmg/ddi048. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
