This is a preprint.
E. coli transcription factors regulate promoter activity by a universal, homeostatic mechanism
- PMID: 39713321
- PMCID: PMC11661191
- DOI: 10.1101/2024.12.09.627516
E. coli transcription factors regulate promoter activity by a universal, homeostatic mechanism
Abstract
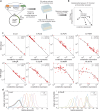
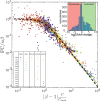
Transcription factors (TFs) may activate or repress gene expression through an interplay of different mechanisms, including RNA polymerase (RNAP) recruitment, exclusion, and initiation. TFs often have drastically different regulatory behaviors depending on promoter context and interacting cofactors. However, the detailed mechanisms by which each TF affects transcription and produce promoter-dependent regulation is unclear. Here, we discover that a simple model explains the regulatory effects of E. coli TFs in a range of contexts. Specifically, we measure the relationship between basal promoter activity and its regulation by diverse TFs and find that the contextual changes in TF function are determined entirely by the basal strength of the regulated promoter: TFs exert lower fold-change on stronger promoters under a precise inverse scaling. Remarkably, this scaling relationship holds for both activators and repressors, indicating a universal mechanism of gene regulation. Our data, which spans between 100-fold activation to 1000-fold repression, is consistent with a model of regulation driven by stabilization of RNAP at the promoter for every TF. Crucially, this indicates that TFs naturally act to maintain homeostatic expression levels across genetic or environmental perturbations, ensuring robust expression of regulated genes.
Conflict of interest statement
Competing interests: There are no competing interests to declare.
Figures



Similar articles
-
A systematic survey of TF function in E. coli suggests RNAP stabilization is a prevalent strategy for both repressors and activators.Nucleic Acids Res. 2025 Feb 8;53(4):gkaf058. doi: 10.1093/nar/gkaf058. Nucleic Acids Res. 2025. PMID: 39921566 Free PMC article.
-
Quantifying the regulatory role of individual transcription factors in Escherichia coli.Cell Rep. 2021 Nov 9;37(6):109952. doi: 10.1016/j.celrep.2021.109952. Cell Rep. 2021. PMID: 34758318 Free PMC article.
-
Screening of promoter-specific transcription factors: multiple regulators for the sdiA gene involved in cell division control and quorum sensing.Microbiology (Reading). 2013 Dec;159(Pt 12):2501-2512. doi: 10.1099/mic.0.067538-0. Epub 2013 Sep 11. Microbiology (Reading). 2013. PMID: 24025606
-
What do Transcription Factors Interact With?J Mol Biol. 2021 Jul 9;433(14):166883. doi: 10.1016/j.jmb.2021.166883. Epub 2021 Feb 20. J Mol Biol. 2021. PMID: 33621520 Free PMC article. Review.
-
The σ Subunit-Remodeling Factors: An Emerging Paradigms of Transcription Regulation.Front Microbiol. 2020 Jul 29;11:1798. doi: 10.3389/fmicb.2020.01798. eCollection 2020. Front Microbiol. 2020. PMID: 32849409 Free PMC article. Review.
References
-
- Record M. T. Jr., Reznikoff W., Craig M., McQuade K., Schlax P., Escherichia coli RNA polymerase (sigma70) promoters and the kinetics of the steps of transcription initiation, in In Escherichia coli and Salmonella Cellular and Molecular Biology (ASM Press, Washington DC: ), pp. 792–821 (1996).
-
- Zhang F., Carothers J. M., Keasling J. D., Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nature Biotechnolgy. 30 (4), 354–359 (2012). - PubMed
-
- Barnard A., Wolfe A., Busby S., Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Current Opinion in Microbiology. 7, 102–108 (2004). - PubMed
-
- Müller J., Oehler S., Müller-Hill B., Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. Journal of Molecular Biology. 257 (1), 21–9 (1996). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
