This is a preprint.
Transcription factors form a ternary complex with NIPBL/MAU2 to localize cohesin at enhancers
- PMID: 39713324
- PMCID: PMC11661173
- DOI: 10.1101/2024.12.09.627537
Transcription factors form a ternary complex with NIPBL/MAU2 to localize cohesin at enhancers
Update in
-
Transcription factors form a ternary complex with NIPBL/MAU2 to localize cohesin at enhancers.Nucleic Acids Res. 2025 May 10;53(9):gkaf415. doi: 10.1093/nar/gkaf415. Nucleic Acids Res. 2025. PMID: 40377219 Free PMC article.
Abstract
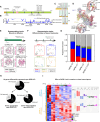
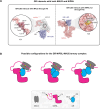
While the cohesin complex is a key player in genome architecture, how it localizes to specific chromatin sites is not understood. Recently, we and others have proposed that direct interactions with transcription factors lead to the localization of the cohesin-loader complex (NIPBL/MAU2) within enhancers. Here, we identify two clusters of LxxLL motifs within the NIPBL sequence that regulate NIPBL dynamics, interactome, and NIPBL-dependent transcriptional programs. One of these clusters interacts with MAU2 and is necessary for the maintenance of the NIPBL-MAU2 heterodimer. The second cluster binds specifically to the ligand-binding domains of steroid receptors. For the glucocorticoid receptor (GR), we examine in detail its interaction surfaces with NIPBL and MAU2. Using AlphaFold2 and molecular docking algorithms, we uncover a GR-NIPBL-MAU2 ternary complex and describe its importance in GR-dependent gene regulation. Finally, we show that multiple transcription factors interact with NIPBL-MAU2, likely using interfaces other than those characterized for GR.
Conflict of interest statement
CONFLICT OF INTEREST The authors declare no competing interests.
Figures



Similar articles
-
Transcription factors form a ternary complex with NIPBL/MAU2 to localize cohesin at enhancers.Nucleic Acids Res. 2025 May 10;53(9):gkaf415. doi: 10.1093/nar/gkaf415. Nucleic Acids Res. 2025. PMID: 40377219 Free PMC article.
-
MAU2 and NIPBL Variants Impair the Heterodimerization of the Cohesin Loader Subunits and Cause Cornelia de Lange Syndrome.Cell Rep. 2020 May 19;31(7):107647. doi: 10.1016/j.celrep.2020.107647. Cell Rep. 2020. PMID: 32433956
-
Localisation of the SMC loading complex Nipbl/Mau2 during mammalian meiotic prophase I.Chromosoma. 2014 Jun;123(3):239-52. doi: 10.1007/s00412-013-0444-7. Epub 2013 Nov 28. Chromosoma. 2014. PMID: 24287868 Free PMC article.
-
Roles of NIPBL in maintenance of genome stability.Semin Cell Dev Biol. 2019 Jun;90:181-186. doi: 10.1016/j.semcdb.2018.08.005. Epub 2018 Aug 17. Semin Cell Dev Biol. 2019. PMID: 30096364 Review.
-
NIPBL and cohesin: new take on a classic tale.Trends Cell Biol. 2023 Oct;33(10):860-871. doi: 10.1016/j.tcb.2023.03.006. Epub 2023 Apr 15. Trends Cell Biol. 2023. PMID: 37062615 Review.
References
-
- Sanborn A.L., Rao S.S., Huang S.C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J. et al. (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A, 112, E6456–6465. - PMC - PubMed
-
- Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A. and Nasmyth K. (2000) Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell, 5, 243–254. - PubMed
-
- Alonso-Gil D. and Losada A. (2023) NIPBL and cohesin: new take on a classic tale. Trends Cell Biol, 33, 860–871. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
