This is a preprint.
Stable isotope fingerprinting can directly link intestinal microorganisms with their carbon source and captures diet-induced substrate switching in vivo
- PMID: 39713332
- PMCID: PMC11661160
- DOI: 10.1101/2024.12.10.627769
Stable isotope fingerprinting can directly link intestinal microorganisms with their carbon source and captures diet-induced substrate switching in vivo
Update in
-
Metaproteomics-based stable isotope fingerprinting links intestinal bacteria to their carbon source and captures diet-induced substrate switching.ISME J. 2025 Jan 2;19(1):wraf127. doi: 10.1093/ismejo/wraf127. ISME J. 2025. PMID: 40581743 Free PMC article.
Abstract
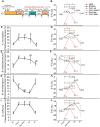
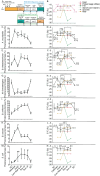
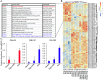
Diet has strong impacts on the composition and function of the gut microbiota with implications for host health. Therefore, it is critical to identify the dietary components that support growth of specific microorganisms in vivo. We used protein-based stable isotope fingerprinting (Protein-SIF) to link microbial species in gut microbiota to their carbon sources by measuring each microbe's natural 13C content (δ13C) and matching it to the 13C content of available substrates. We fed gnotobiotic mice, inoculated with a 13 member microbiota, diets in which the 13C content of all components was known. We varied the source of protein, fiber or fat to observe 13C signature changes in microbial consumers of these substrates. We observed significant changes in the δ13C values and abundances of specific microbiota species, as well as host proteins, in response to changes in 13C signature or type of protein, fiber, and fat sources. Using this approach we were able to show that upon switching dietary source of protein, fiber, or fat (1) some microbial species continued to obtain their carbon from the same dietary component (e.g., protein); (2) some species switched their main substrate type (e.g., from protein to carbohydrates); and (3) some species might derive their carbon through foraging on host compounds. Our results demonstrate that Protein-SIF can be used to identify the dietary-derived substrates assimilated into proteins by microbes in the intestinal tract; this approach holds promise for the analysis of microbiome substrate usage in humans without the need of substrate labeling.
Significance: The gut microbiota plays a critical role in the health of animals including humans, influencing metabolism, the immune system, and even behavior. Diet is one of the most significant factors in determining the function and composition of the gut microbiota, but our understanding of how specific dietary components directly impact individual microbes remains limited. We present the application of an approach that measures the carbon isotope "fingerprint" of proteins in biological samples. This fingerprint is similar to the fingerprint of the substrate used to make the proteins. We describe how we used this approach in mice to determine which dietary components specific intestinal microbes use as carbon sources to make their proteins. This approach can directly identify components of an animal's diet that are consumed by gut microbes.
Keywords: Gut microbiota; metaproteomics; microbiome; protein-SIF.
Figures

Similar articles
-
Metaproteomics-based stable isotope fingerprinting links intestinal bacteria to their carbon source and captures diet-induced substrate switching.ISME J. 2025 Jan 2;19(1):wraf127. doi: 10.1093/ismejo/wraf127. ISME J. 2025. PMID: 40581743 Free PMC article.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
References
-
- Perler B. K., Friedman E. S., Wu G. D., The Role of the Gut Microbiota in the Relationship Between Diet and Human Health. Annu. Rev. Physiol. 85, 449–468 (2023). - PubMed
-
- Shimotoyodome A., Meguro S., Hase T., Tokimitsu I., Sakata T., Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 125, 525–531 (2000). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
