This is a preprint.
An endogenous cluster of target-directed microRNA degradation sites induces decay of distinct microRNA families
- PMID: 39713366
- PMCID: PMC11661237
- DOI: 10.1101/2024.12.11.627053
An endogenous cluster of target-directed microRNA degradation sites induces decay of distinct microRNA families
Abstract
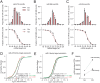
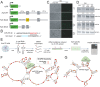
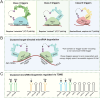
While much is known about miRNA biogenesis and canonical miRNA targeting, relatively less is understood about miRNA decay. The major miRNA decay pathway in metazoans is mediated through target-directed miRNA degradation (TDMD), in which certain RNAs can "trigger" miRNA decay. All known triggers for TDMD base pair with the miRNA seed, and extensively base pair on the miRNA 3' end, a pattern that supposedly induces a TDMD-competent conformational change of Argonaute (Ago), allowing for miRNA turnover. Here, we utilized Ago1-CLASH to find that the Drosophila transcript Kah contains at least two triggers, a "trigger cluster", against miR-9b and the miR-279 family. One of these triggers contains minimal/non-canonical 3' end base pairing but is still sufficient to induce TDMD of the entire miR-279 family. We found that these clustered triggers likely lack cooperativity, the minimal 3' pairing is required for miR-279 family turnover, and probed the in-cell RNA structure of the Kah trigger cluster. Overall, this study expands the list of endogenous triggers and the unexpectedly complex regulatory network governing miRNA degradation.
Keywords: AGO-CLASH; SHAPE; TDMD; microRNA.
Figures



Similar articles
-
Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins.Genes Dev. 2021 Dec 1;35(23-24):1595-1609. doi: 10.1101/gad.348874.121. Epub 2021 Nov 24. Genes Dev. 2021. PMID: 34819352 Free PMC article.
-
Screening of Drosophila microRNA-degradation sequences reveals Argonaute1 mRNA's role in regulating miR-999.Nat Commun. 2023 Apr 13;14(1):2108. doi: 10.1038/s41467-023-37819-9. Nat Commun. 2023. PMID: 37055443 Free PMC article.
-
Target-directed microRNA degradation: Mechanisms, significance, and functional implications.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1832. doi: 10.1002/wrna.1832. Wiley Interdiscip Rev RNA. 2024. PMID: 38448799 Free PMC article. Review.
-
CLASHub: an integrated database and analytical platform for microRNA-target interactions.bioRxiv [Preprint]. 2025 Aug 7:2025.08.05.668543. doi: 10.1101/2025.08.05.668543. bioRxiv. 2025. PMID: 40799581 Free PMC article. Preprint.
-
To kill a microRNA: emerging concepts in target-directed microRNA degradation.Nucleic Acids Res. 2024 Feb 28;52(4):1558-1574. doi: 10.1093/nar/gkae003. Nucleic Acids Res. 2024. PMID: 38224449 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
