This is a preprint.
Long-read Transcriptomics of Caviid Gammaherpesvirus 1: Compiling a Comprehensive RNA Atlas
- PMID: 39713410
- PMCID: PMC11661164
- DOI: 10.1101/2024.12.11.627975
Long-read Transcriptomics of Caviid Gammaherpesvirus 1: Compiling a Comprehensive RNA Atlas
Update in
-
Long-read transcriptomics of caviid gammaherpesvirus 1: compiling a comprehensive RNA atlas.mSystems. 2025 Mar 18;10(3):e0167824. doi: 10.1128/msystems.01678-24. Epub 2025 Feb 27. mSystems. 2025. PMID: 40013795 Free PMC article.
Abstract
Caviid gammaherpesvirus 1 (CaGHV-1), formerly known as the guinea pig herpes-like virus, is an oncogenic gammaherpesvirus with a sequenced genome but an as-yet uncharacterized transcriptome. Using nanopore long-read RNA sequencing, we annotated the CaGHV-1 genome and constructed a detailed transcriptomic atlas. Our findings reveal diverse viral mRNAs and non-coding RNAs, along with mapped promoter elements for each viral gene. We demonstrated that the CaGHV-1 RTA lytic cycle transcription factor activates its own promoter, similar to KSHV, and that the CaGHV-1 ORF50 promoter responds to RTA proteins from other gammaherpesviruses, highlighting the evolutionary conservation of RTA-mediated transcriptional mechanisms. Additionally, our analysis uncovered extensive transcriptional overlap within the viral genome, suggesting a role in regulating global gene expression. Given its tumorigenic properties, broad host range, and non-human pathogenicity, this work establishes CaGHV-1 as a promising small animal model for investigating human gammaherpesvirus pathogenesis.
Keywords: Caviid gammaherpesvirus 1 (CaGHV-1); direct RNA sequencing; gammaherpesvirus; guinea pig herpes-like virus; long-read sequencing; luciferase reporter assay; nanopore sequencing; transcriptome.
Conflict of interest statement
Conflicts of interests The authors do not declare any conflicts of interest.
Figures
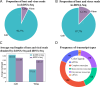


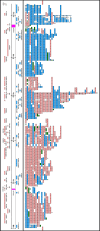


Similar articles
-
Long-read transcriptomics of caviid gammaherpesvirus 1: compiling a comprehensive RNA atlas.mSystems. 2025 Mar 18;10(3):e0167824. doi: 10.1128/msystems.01678-24. Epub 2025 Feb 27. mSystems. 2025. PMID: 40013795 Free PMC article.
-
A new era in gammaherpesvirus transcriptomics: high-resolution profiling and model development.mSystems. 2025 Jul 23:e0020825. doi: 10.1128/msystems.00208-25. Online ahead of print. mSystems. 2025. PMID: 40698955
-
Guinea pig herpes like virus is a gamma herpesvirus.Virus Genes. 2024 Apr;60(2):148-158. doi: 10.1007/s11262-024-02054-x. Epub 2024 Feb 10. Virus Genes. 2024. PMID: 38340271 Free PMC article.
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
-
PAN RNA: transcriptional exhaust from a viral engine.J Biomed Sci. 2020 Mar 7;27(1):41. doi: 10.1186/s12929-020-00637-y. J Biomed Sci. 2020. PMID: 32143650 Free PMC article. Review.
References
-
- Kieff E RA. 2007. Epstein-Barr Virus and its replication. Fields Virol 5th:2603–2654.
-
- Rajcáni J, Blaskovic D, Svobodová J, Ciampor F, Hucková D, Staneková D. 1985. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol 29:51–60. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
