This is a preprint.
Structural serology of polyclonal antibody responses to mRNA-1273 and NVX-CoV2373 COVID-19 vaccines
- PMID: 39713412
- PMCID: PMC11661243
- DOI: 10.1101/2024.12.11.628030
Structural serology of polyclonal antibody responses to mRNA-1273 and NVX-CoV2373 COVID-19 vaccines
Update in
-
Structural serology of polyclonal antibody responses to mRNA-1273 and NVX-CoV2373 COVID-19 vaccines.Cell Rep. 2025 Jul 22;44(7):115986. doi: 10.1016/j.celrep.2025.115986. Epub 2025 Jul 8. Cell Rep. 2025. PMID: 40632654 Free PMC article. Clinical Trial.
Abstract
Current COVID-19 vaccines are largely limited in their ability to induce broad, durable immunity against emerging viral variants. Design and development of improved vaccines utilizing existing platforms requires an in-depth understanding of the antigenic and immunogenic properties of available vaccines. Here we examined the antigenicity of two of the original COVID-19 vaccines, mRNA-1273 and NVX-CoV2373, by electron microscopy-based polyclonal epitope mapping (EMPEM) of serum from immunized non-human primates (NHPs) and clinical trial donors. Both vaccines induce diverse polyclonal antibody (pAb) responses to the N-terminal domain (NTD) in addition to the receptor-binding domain (RBD) of the Spike protein, with the NTD supersite being an immunodominant epitope. High-resolution cryo-EMPEM studies revealed extensive pAb responses to and around the supersite with unique angles of approach and engagement. NTD supersite pAbs were also the most susceptible to variant mutations compared to other specificities, indicating that ongoing Spike ectodomain-based vaccine design strategies should consider immuno-masking this site to prevent induction of these strain-specific responses.
Conflict of interest statement
Declaration of interests: KSC, BSG, and ABW are inventors on a US Patent No. 10/960,070 B2 entitled “Prefusion Coronavirus Spike Proteins and Their Use.” KSC and BSG are inventors on US Patent Application No. 202117798021 entitled “SARS-CoV-2 Vaccine.” ABW is an inventor on a patent US patent 11217328 entitled “Epitope Mapping Method.” BG, RD and DKE are employees of Moderna, Inc. and may hold stock/stock options in the company. AR, MG-X, NP, and GS are employees of Novavax, Inc. and hold stock or stock options. All other authors have no competing interests to declare.
Figures



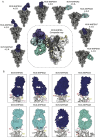



References
-
- Harder T., Koch J., Vygen-Bonnet S., Kulper-Schiek W., Pilic A., Reda S., Scholz S., and Wichmann O. (2021). Efficacy and effectiveness of COVID-19 vaccines against SARS-CoV-2 infection: interim results of a living systematic review, 1 January to 14 May 2021. Eurosurveillance 26, 2100563. 10.2807/1560-7917.es.2021.26.28.2100563. - DOI - PMC - PubMed
-
- Lau J.J., Cheng S.M.S., Leung K., Lee C.K., Hachim A., Tsang L.C.H., Yam K.W.H., Chaothai S., Kwan K.K.H., Chai Z.Y.H., et al. (2023). Real-world COVID-19 vaccine effectiveness against the Omicron BA.2 variant in a SARS-CoV-2 infection-naive population. Nat. Med. 29, 348–357. 10.1038/s41591-023-02219-5. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
