This is a preprint.
Calmodulin binding is required for calcium mediated TRPA1 desensitization
- PMID: 39713425
- PMCID: PMC11661184
- DOI: 10.1101/2024.12.11.627969
Calmodulin binding is required for calcium mediated TRPA1 desensitization
Abstract
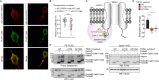
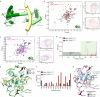
Calcium (Ca2+) ions affect nearly all aspects of biology. Excessive Ca2+ entry is cytotoxic and Ca2+-mobilizing receptors have evolved diverse mechanisms for tight regulation that often include Calmodulin (CaM). TRPA1, an essential Ca2+-permeable ion channel involved in pain signaling and inflammation, exhibits complex Ca2+ regulation with initial channel potentiation followed by rapid desensitization. The molecular mechanisms of TRPA1 Ca2+ regulation and whether CaM plays a role remain elusive. We find that TRPA1 binds CaM best at basal Ca2+ concentration, that they co-localize in resting cells, and that CaM suppresses TRPA1 activity. Combining biochemical, biophysical, modeling, NMR spectroscopy, and functional approaches, we identify an evolutionarily conserved, high-affinity CaM binding element in the distal TRPA1 C-terminus (DCTCaMBE). Genetic or biochemical perturbation of Ca2+/CaM binding to the TRPA1 DCTCaMBE yields hyperactive channels that exhibit drastic slowing of desensitization with no effect on potentiation. Ca2+/CaM TRPA1 regulation does not require the N-lobe, raising the possibility that CaM is not the Ca2+ sensor, per se. Higher extracellular Ca2+ can partially rescue slowed desensitization suggesting Ca2+/CaM binding to the TRPA1 DCTCaMBE primes an intrinsic TRPA1 Ca2+ binding site that, upon binding Ca2+, triggers rapid desensitization. Collectively, our results identify a critical regulatory element in an unstructured TRPA1 region highlighting the importance of these domains, they reveal Ca2+/CaM is an essential TRPA1 auxiliary subunit required for rapid desensitization that establishes proper channel function with implications for all future TRPA1 work, and they uncover a mechanism for receptor regulation by Ca2+/CaM that expands the scope of CaM biology.
Conflict of interest statement
Competing Interests: The authors declare no competing interests.
Figures






Similar articles
-
Calmodulin is responsible for Ca2+-dependent regulation of TRPA1 Channels.Sci Rep. 2017 Mar 23;7:45098. doi: 10.1038/srep45098. Sci Rep. 2017. PMID: 28332600 Free PMC article.
-
Distinct properties of Ca2+-calmodulin binding to N- and C-terminal regulatory regions of the TRPV1 channel.J Gen Physiol. 2012 Nov;140(5):541-55. doi: 10.1085/jgp.201210810. J Gen Physiol. 2012. PMID: 23109716 Free PMC article.
-
CaV channels reject signaling from a second CaM in eliciting Ca2+-dependent feedback regulation.J Biol Chem. 2020 Oct 30;295(44):14948-14962. doi: 10.1074/jbc.RA120.013777. Epub 2020 Aug 20. J Biol Chem. 2020. PMID: 32820053 Free PMC article.
-
L-Type Ca2+ Channel Regulation by Calmodulin and CaBP1.Biomolecules. 2021 Dec 2;11(12):1811. doi: 10.3390/biom11121811. Biomolecules. 2021. PMID: 34944455 Free PMC article. Review.
-
Regulation of Connexin Gap Junctions and Hemichannels by Calcium and Calcium Binding Protein Calmodulin.Int J Mol Sci. 2020 Nov 2;21(21):8194. doi: 10.3390/ijms21218194. Int J Mol Sci. 2020. PMID: 33147690 Free PMC article. Review.
References
-
- Clapham D. E. Calcium signaling. Cell 131, 1047–1058 (2007). - PubMed
-
- Averbeck B. in Encyclopedia of Pain 1320–1322 (Springer, 2007).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
