Single-Cell Sequencing Combined with Transcriptome Sequencing to Explore the Molecular Mechanisms Related to Skin Photoaging
- PMID: 39713718
- PMCID: PMC11662644
- DOI: 10.2147/JIR.S496328
Single-Cell Sequencing Combined with Transcriptome Sequencing to Explore the Molecular Mechanisms Related to Skin Photoaging
Abstract
Background: The aging of skin is a diversified biological phenomenon, influenced by a combination of genetic and environmental factors. However, the specific mechanism of skin photoaging is not yet completely elucidated.
Methods: Gene expression profiles for photoaging patients were obtained from the Gene Expression Omnibus (GEO) collection. We conducted single-cell and intercellular communication investigations to identify potential gene sets. Predictive models were created using LASSO regression. The relationships between genes and immune cells were investigated using single sample gene set enrichment analysis (ssGSEA) and gene set variance analysis (GSVA). The molecular processes of important genes were studied using gene enrichment analysis. A miRNA network was created to look for target miRNAs connected with important genes, and transcriptional regulation analysis was used to identify related transcription factors. Finally, merging gene co-expression networks with drug prediction shows molecular pathways of photoaging and potential treatment targets. Furthermore, we validated the role of key genes, immune cell infiltration, and the Adenosine 5'-monophosphate (AMP)-activated protein kinase (AMPK) pathway in photoaging, which were identified through bioinformatics analysis, using in vivo reverse transcription quantitative PCR (RT-qPCR), immunofluorescence labeling, and Western blotting.
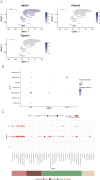
Results: This study discovered three key genes, including Atp2b1, Plekho2, and Tspan13, which perform crucial functions in the photoaging process. Immune cell infiltration analysis showed increased M1 macrophages and CD4 memory T cells in the photoaging group. Further signaling pathway analysis indicated that these key genes are enriched in multiple immune and metabolic pathways. The significant roles of Atp2b1, Plekho2, Tspan13, M1 macrophages infiltration, CD4 memory T cells infiltration and the AMPK pathway in photoaging was validated in vivo.
Conclusion: This research revealed the underlying molecular mechanisms of photoaging, indicating that key genes such as Atp2b1 and Tspan13 play crucial roles in the regulation of immune cell infiltration and metabolic pathways. These findings provide a new theory for the treatment of photoaging and provide prospective targets for the advancement of relevant drugs.
Keywords: AMPK pathway; Atp2b1; Plekho2; Tspan13; immune cell infiltration; skin photoaging.
© 2024 Hu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Biomarkers of UVB radiation-related senescent fibroblasts.Sci Rep. 2024 Jan 9;14(1):933. doi: 10.1038/s41598-023-51058-4. Sci Rep. 2024. PMID: 38195709 Free PMC article.
-
Bioinformatics-Based Identification of Key Prognostic Genes in Neuroblastoma with a Focus on Immune Cell Infiltration and Diagnostic Potential of VGF.Pharmgenomics Pers Med. 2024 Oct 10;17:453-472. doi: 10.2147/PGPM.S461072. eCollection 2024. Pharmgenomics Pers Med. 2024. PMID: 39403102 Free PMC article.
-
Gene expression analysis in endometriosis: Immunopathology insights, transcription factors and therapeutic targets.Front Immunol. 2022 Nov 30;13:1037504. doi: 10.3389/fimmu.2022.1037504. eCollection 2022. Front Immunol. 2022. PMID: 36532015 Free PMC article.
-
[Exploration of key ferroptosis-related genes as therapeutic targets for sepsis based on bioinformatics and the depiction of their immune profiles characterization].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2024 Oct;36(10):1025-1032. doi: 10.3760/cma.j.cn121430-20240524-00457. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2024. PMID: 39586719 Chinese.
-
Multi-omics analysis to explore the molecular mechanisms related to keloid.Burns. 2025 Apr;51(3):107396. doi: 10.1016/j.burns.2025.107396. Epub 2025 Jan 21. Burns. 2025. PMID: 39874886
References
LinkOut - more resources
Full Text Sources
Research Materials

