Millisecond Label-Free Single Peptide Detection and Identification Using Nanoscale Electrochromatography and Resistive Pulse Sensing
- PMID: 39713813
- PMCID: PMC12006914
- DOI: 10.1021/acs.analchem.4c04542
Millisecond Label-Free Single Peptide Detection and Identification Using Nanoscale Electrochromatography and Resistive Pulse Sensing
Abstract
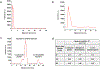
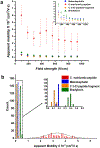
We are developing a unique protein identification method that consists of generating peptides proteolytically from a single protein molecule (i.e., peptide fingerprints) with peptide detection and identification carried out using nanoscale electrochromatography and label-free resistive pulse sensing (RPS). As a step in realizing this technology, we report herein the nanoscale electrochromatography of model peptides using thermoplastic columns with surfaces engineered to identify peptides via their molecularly dependent mobility (i.e., time-of-flight, ToF). ToFs were elucidated using a dual in-plane nanopore sensor, which consisted of two in-plane nanopores placed on either end of the nanoelectrochromatography column. The surface of the nanocolumn, which consisted of poly(methyl methacrylate) (PMMA), was activated with an O2 plasma, creating surface carboxylic acid groups (-COOH) inducing a surface charge on the column wall as well as affecting its hydrophilicity. To understand scaling effects, we carried out microchip and nanochannel electrochromatography of the peptides labeled with an ATTO 532 reporter to allow for single-molecule tracking. Our results indicated that the apparent mobilities of the model peptides did not allow for their separation in a microchannel, but when performed in a nanocolumn, clear differences in their apparent mobilities could be observed especially when operated at high electric field strengths. We next performed label-free detection of peptides using the dual in-plane nanopore sensor with the two pores separated by a 5 μm (length) column with a 50 nm width and depth. When a single peptide molecule passed through an in-plane nanopore, the sensor read a pair of resistive pulses with a time difference equivalent to ToF. We identified the peptides by evaluating their ToF, normalized RPS current transient amplitude (ΔI/I0), and RPS peak dwell time (td). We could identify the model peptides with nearly 100% classification accuracy at the single-molecule level using machine learning with a single molecule measurement requiring <10 ms.
Figures



Similar articles
-
Detection and identification of single ribonucleotide monophosphates using a dual in-plane nanopore sensor made in a thermoplastic via replication.Lab Chip. 2024 May 14;24(10):2721-2735. doi: 10.1039/d3lc01062g. Lab Chip. 2024. PMID: 38656267 Free PMC article.
-
Label-Free Identification of Single Mononucleotides by Nanoscale Electrophoresis.Small. 2021 Oct;17(42):e2102567. doi: 10.1002/smll.202102567. Epub 2021 Sep 23. Small. 2021. PMID: 34558175 Free PMC article.
-
In-Plane, In-Series Nanopores with Circular Cross Sections for Resistive-Pulse Sensing.ACS Nano. 2022 May 24;16(5):7352-7360. doi: 10.1021/acsnano.1c08680. Epub 2022 May 2. ACS Nano. 2022. PMID: 35500295 Free PMC article.
-
The Utility of Nanopore Technology for Protein and Peptide Sensing.Proteomics. 2018 Sep;18(18):e1800026. doi: 10.1002/pmic.201800026. Epub 2018 Aug 5. Proteomics. 2018. PMID: 29952121 Free PMC article. Review.
-
Separation, detection and quantitation of peptides by liquid chromatography and capillary electrochromatography.J Chromatogr A. 2009 Mar 6;1216(10):1825-37. doi: 10.1016/j.chroma.2008.12.052. Epub 2008 Dec 25. J Chromatogr A. 2009. PMID: 19131068 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

