Impairment of lipid homeostasis causes lysosomal accumulation of endogenous protein aggregates through ESCRT disruption
- PMID: 39713930
- PMCID: PMC11666243
- DOI: 10.7554/eLife.86194
Impairment of lipid homeostasis causes lysosomal accumulation of endogenous protein aggregates through ESCRT disruption
Abstract
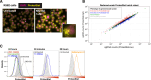
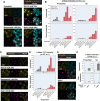
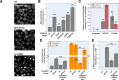
Protein aggregation increases during aging and is a pathological hallmark of many age-related diseases. Protein homeostasis (proteostasis) depends on a core network of factors directly influencing protein production, folding, trafficking, and degradation. Cellular proteostasis also depends on the overall composition of the proteome and numerous environmental variables. Modulating this cellular proteostasis state can influence the stability of multiple endogenous proteins, yet the factors contributing to this state remain incompletely characterized. Here, we performed genome-wide CRISPRi screens to elucidate the modulators of proteostasis state in mammalian cells, using a fluorescent dye to monitor endogenous protein aggregation. These screens identified known components of the proteostasis network and uncovered a novel link between protein and lipid homeostasis. Increasing lipid uptake and/or disrupting lipid metabolism promotes the accumulation of sphingomyelins and cholesterol esters and drives the formation of detergent-insoluble protein aggregates at the lysosome. Proteome profiling of lysosomes revealed ESCRT accumulation, suggesting disruption of ESCRT disassembly, lysosomal membrane repair, and microautophagy. Lipid dysregulation leads to lysosomal membrane permeabilization but does not otherwise impact fundamental aspects of lysosomal and proteasomal functions. Together, these results demonstrate that lipid dysregulation disrupts ESCRT function and impairs proteostasis.
Keywords: CRISPR; ESCRT; aggregation; cell biology; human; lipid dysregulation; lysosome; proteostasis.
© 2023, Yong et al.
Conflict of interest statement
JY, JV, NV, NO, ML, JL, KH, FM, BB, CJ Employee of Calico Life Sciences LLC, MK Former employee of Calico Life Sciences LLC
Figures











Update of
- doi: 10.1101/2022.11.23.517579
- doi: 10.7554/eLife.86194.1
- doi: 10.7554/eLife.86194.2
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

