SCovid v2.0: a comprehensive resource to decipher the molecular characteristics across tissues in COVID-19 and other human coronaviruses
- PMID: 39714149
- PMCID: PMC11792472
- DOI: 10.1128/spectrum.01933-24
SCovid v2.0: a comprehensive resource to decipher the molecular characteristics across tissues in COVID-19 and other human coronaviruses
Abstract
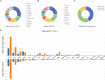
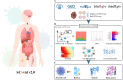
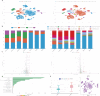
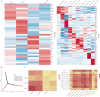
SCovid v2.0 (http://bio-annotation.cn/scovid or http://bio-computing.hrbmu.edu.cn/scovid/) is an updated database designed to assist researchers in uncovering the molecular characteristics of coronavirus disease 2019 (COVID-19) across various tissues through transcriptome sequencing. Compared with its predecessor, SCovid v2.0 is enhanced with comprehensive data, practical functionalities, and a reconstructed pipeline. The current release includes (i) 3,544,360 cells from 45 single-cell RNA-seq (scRNA-seq) data sets encompassing 789 samples from 15 tissues; (ii) the addition of 62 COVID-19 bulk RNA-seq data comprising 1,688 samples from 12 tissues; (iii) incorporation of seven bulk RNA-seq data sets related to other human coronaviruses, such as HCoV-229E, HCoV-OC43, and MERS-CoV for a thorough comparative analysis of pan-coronavirus mechanisms in COVID-19; and (iv) systematic comparisons between the data sets conducted using standardized procedures. Furthermore, we have developed an advanced search engine and upgraded web interface to browse, search, visualize, and download detailed information. Overall, SCovid v2.0 is a valuable resource for exploring molecular characteristics of COVID-19 across different tissues.
Importance: This manuscript provides a comprehensive analysis of the molecular characteristics of COVID-19 through cross-tissue transcriptome analysis, contributing to the understanding of COVID-19 by clinicians and scientists. Considering the cyclical nature of coronavirus outbreaks, this updated database adds transcriptome data on other human coronaviruses, contributing to potential and existing mechanisms of other human coronaviruses.
Keywords: COVID-19; bulk RNA-seq; molecular characteristics; other human coronaviruses; single-cell RNA-seq.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017 - DOI - PMC - PubMed
-
- Edahiro R, Shirai Y, Takeshima Y, Sakakibara S, Yamaguchi Y, Murakami T, Morita T, Kato Y, Liu Y-C, Motooka D, et al. 2023. Single-cell analyses and host genetics highlight the role of innate immune cells in COVID-19 severity. Nat Genet 55:753–767. doi: 10.1038/s41588-023-01375-1 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

