Henipaviruses: epidemiology, ecology, disease, and the development of vaccines and therapeutics
- PMID: 39714175
- PMCID: PMC11905374
- DOI: 10.1128/cmr.00128-23
Henipaviruses: epidemiology, ecology, disease, and the development of vaccines and therapeutics
Abstract
SUMMARYHenipaviruses were first identified 30 years ago and have since been associated with over 30 outbreaks of disease in humans. Highly pathogenic henipaviruses include Hendra virus (HeV) and Nipah virus (NiV), classified as biosafety level 4 pathogens. In addition, NiV has been listed as a priority pathogen by the World Health Organization (WHO), the Coalition for Epidemic Preparedness Innovations (CEPI), and the UK Vaccines Research and Development Network (UKVN). Here, we re-examine epidemiological, ecological, clinical, and pathobiological studies of HeV and NiV to provide a comprehensive guide of the current knowledge and application to identify and evaluate countermeasures. We also discuss therapeutic and vaccine development efforts. Furthermore, with case identification, prevention, and treatment in mind, we highlight limitations in research and recognize gaps necessitating additional studies.
Keywords: Hendra virus; Nipah virus; animal model; antiviral; clinical disease; epidemiology; henipavirus; therapeutic; vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
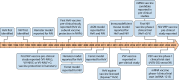
Figures


References
-
- Lamb RA, Parks GD. 2013. Paramyxoviridae: the viruses and their replicationp 957–995. In Fields BN, Knipe DM, Howley PM (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA, USA.
-
- Madera S, Kistler A, Ranaivoson HC, Ahyong V, Andrianiaina A, Andry S, Raharinosy V, Randriambolamanantsoa TH, Ravelomanantsoa NAF, Tato CM, DeRisi JL, Aguilar HC, Lacoste V, Dussart P, Heraud J-M, Brook CE. 2022. Discovery and genomic characterization of a novel Henipavirus, Angavokely virus, from fruit bats in Madagascar. J Virol 96:e0092122. doi: 10.1128/jvi.00921-22 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

