Patterns of spontaneous and induced genomic alterations in Yarrowia lipolytica
- PMID: 39714191
- PMCID: PMC11784153
- DOI: 10.1128/aem.01678-24
Patterns of spontaneous and induced genomic alterations in Yarrowia lipolytica
Abstract
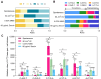
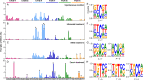
This study explored the genomic alterations in Yarrowia lipolytica, a key yeast in industrial biotechnology, under both spontaneous and mutagen-induced conditions. Our findings reveal that spontaneous mutations occur at a rate of approximately 4 × 10-10 events per base pair per cell division, primarily manifesting as single-nucleotide variations (SNVs) and small insertions and deletions (InDels). Notably, C-to-T/G-to-A transitions and C-to-A/G-to-T transversions dominate the spontaneous SNVs, while 1 bp deletions, likely resulting from template slippage, are the most frequent InDels. Furthermore, chromosomal aneuploidy and rearrangements occur, albeit at a lower frequency. Exposure to ultraviolet (UV) light, methylmethane sulfonate (MMS), and Zeocin significantly enhances the rates of SNVs and alters their mutational spectra in distinct patterns. Notably, Zeocin-induced SNVs are predominantly T-to-A and T-to-G substitutions, often occurring within the 5'-TGT*-3' motif (* denotes the mutated base). Additionally, Zeocin exhibits a higher potency in stimulating InDels compared to UV and MMS. Translesion DNA synthesis is implicated as the primary mechanism behind most Zeocin-induced SNVs and some InDels, whereas non-homologous end joining serves as the main pathway for Zeocin-mediated InDels. Intriguingly, the study identifies the gene YALI1_E21053g, encoding a protein kinase, as negatively associated with Zeocin resistance. Overall, our results not only deepened our knowledge about the genome evolution in Y. lipolytica but also provided reference to develop innovative strategies to harness its genetic potential.IMPORTANCEYarrowia lipolytica exhibits high environmental stress tolerance and lipid metabolism capabilities, making it a microorganism with significant industrial application potential. In this study, we investigated the genomic variation and evolutionary patterns of this yeast under both spontaneous and induced mutation conditions. Our results reveal distinctive mutation spectra induced by different mutagenic conditions and elucidate the underlying genetic mechanisms. We further highlight the roles of non-homologous end joining and translesion synthesis pathways in Zeocin-induced mutations, demonstrating that such treatments can rapidly confer drug resistance to the cells. Overall, our research enhances the understanding of how yeast genomes evolve under various conditions and provides guidance for developing more effective mutagenesis and breeding techniques.
Keywords: Yarrowia lipolytica; Zeocin; drug resistance; genomic alterations; mutation spectrum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The roles of NHEJ and TLS pathways in genomic alterations and phenotypic evolution in the yeast Yarrowia lipolytica.Appl Microbiol Biotechnol. 2025 Aug 15;109(1):183. doi: 10.1007/s00253-025-13575-2. Appl Microbiol Biotechnol. 2025. PMID: 40813729 Free PMC article.
-
Genomic alterations of marine yeast Scheffersomyces spartinae under spontaneous and mutagenic conditions.BMC Genomics. 2025 Mar 25;26(1):297. doi: 10.1186/s12864-025-11479-z. BMC Genomics. 2025. PMID: 40133852 Free PMC article.
-
Identifying new targets for improving terpenoid biosynthesis in Yarrowia lipolytica through random genomic cytosine base editing.Microb Cell Fact. 2025 Aug 20;24(1):192. doi: 10.1186/s12934-025-02819-5. Microb Cell Fact. 2025. PMID: 40830463 Free PMC article.
-
Engineering Yarrowia lipolytica for Use in Biotechnological Applications: A Review of Major Achievements and Recent Innovations.Mol Biotechnol. 2018 Aug;60(8):621-635. doi: 10.1007/s12033-018-0093-4. Mol Biotechnol. 2018. PMID: 29943148 Review.
-
Holistic Approaches in Lipid Production by Yarrowia lipolytica.Trends Biotechnol. 2018 Nov;36(11):1157-1170. doi: 10.1016/j.tibtech.2018.06.007. Epub 2018 Jul 11. Trends Biotechnol. 2018. PMID: 30006239 Review.
Cited by
-
The roles of NHEJ and TLS pathways in genomic alterations and phenotypic evolution in the yeast Yarrowia lipolytica.Appl Microbiol Biotechnol. 2025 Aug 15;109(1):183. doi: 10.1007/s00253-025-13575-2. Appl Microbiol Biotechnol. 2025. PMID: 40813729 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

