Positive regulation of a LuxR family protein, MilO, in mildiomycin biosynthesis
- PMID: 39714196
- PMCID: PMC11784345
- DOI: 10.1128/aem.01654-24
Positive regulation of a LuxR family protein, MilO, in mildiomycin biosynthesis
Abstract
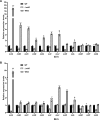
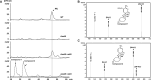
Mildiomycin is a representative peptidyl nucleoside antibiotic and was first isolated from Streptoverticillium rimofaciens, which has been used as an important biological agent to control powdery mildew in plants. Despite its importance, the biosynthetic pathways and regulatory mechanisms remain to be fully elucidated. In this study, we identified MilO as a positive pathway-specific regulator of mildiomycin biosynthesis in the heterologous host Streptomyces avermitilis. Gene disruption of milO resulted in almost loss of mildiomycin production, and it was restored to the level comparable to that in the wild-type strain in complemented strain. Overexpression of milO using host native promoter rpsJp, engineered promotor SP44, and kasOp* led to a 50%, 6.5-fold, and 9.2-fold increase in mildiomycin production compared with the wild-type strain, respectively. Quantitative real-time PCR and electrophoretic mobility shift assay (EMSA) experiments revealed that MilO directly enhances the transcription of the milA gene by 20 folds after 48 h fermentation and indirectly regulates the transcription levels of other genes from milB to milM. Using DNase I footprinting assays, milO was revealed to bind to a 44 bp DNA sequence of the milA promoter region. The binding region consists of three imperfect direct repeats of TGTC(N)3CGGT separated by two-nucleotide spacers and each repeat is important to efficient binding to MilO. In addition, we identified two related compounds by overexpressing milO in a structural gene milN-deficient mutant. Taken together, this study indicates that pathway-specific regulator MilO is essential for mildiomycin biosynthesis and provides an effective strategy to improve the production of mildiomycin and its intermediates.IMPORTANCEAs an important biological agent to control powdery mildew on plants, mildiomycin has been commercialized and used in various plants. However, its regulatory mechanisms and biosynthetic pathways remain unknown. This study provides new insights into the regulation of mildiomycin biosynthesis through MilO, a LuxR family protein that modulates mildiomycin production by directly enhancing the transcription of milA. The yield of mildiomycin was significantly improved by overexpressing milO in a heterologous host. In addition, the positive regulatory effect of milO helped to discover two related compounds, which provide important clues for the timing of uploading of two amino acid side chains during mildiomycin biosynthesis for the first time. In brief, our findings on transcriptional regulation of mildiomycin biosynthesis by milO will be valuable to further increase the yield of mildiomycin and explore its biosynthetic pathways.
Keywords: LuxR regulator; MilO; Streptomyces avermitilis; gene overexpression; mildiomycin; shunt product; yield improvement.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Promoter Engineering Reveals the Importance of Heptameric Direct Repeats for DNA Binding by Streptomyces Antibiotic Regulatory Protein-Large ATP-Binding Regulator of the LuxR Family (SARP-LAL) Regulators in Streptomyces natalensis.Appl Environ Microbiol. 2018 May 1;84(10):e00246-18. doi: 10.1128/AEM.00246-18. Print 2018 May 15. Appl Environ Microbiol. 2018. PMID: 29500267 Free PMC article.
-
Pathway-specific regulation revisited: cross-regulation of multiple disparate gene clusters by PAS-LuxR transcriptional regulators.Appl Microbiol Biotechnol. 2015 Jun;99(12):5123-35. doi: 10.1007/s00253-015-6472-x. Epub 2015 Feb 26. Appl Microbiol Biotechnol. 2015. PMID: 25715784
-
Characterization of a pathway-specific activator of milbemycin biosynthesis and improved milbemycin production by its overexpression in Streptomyces bingchenggensis.Microb Cell Fact. 2016 Sep 7;15(1):152. doi: 10.1186/s12934-016-0552-1. Microb Cell Fact. 2016. PMID: 27604457 Free PMC article.
-
ToyA, a positive pathway-specific regulator for toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628.Appl Microbiol Biotechnol. 2019 Sep;103(17):7071-7084. doi: 10.1007/s00253-019-09959-w. Epub 2019 Jun 29. Appl Microbiol Biotechnol. 2019. PMID: 31256228
-
Positive and negative regulation of GlnR in validamycin A biosynthesis by binding to different loci in promoter region.Appl Microbiol Biotechnol. 2015 Jun;99(11):4771-83. doi: 10.1007/s00253-015-6437-0. Epub 2015 Feb 12. Appl Microbiol Biotechnol. 2015. PMID: 25672849
References
-
- Harada S, Mizuta E, Kishi T. 1981. Structure of mildiomycin, a new antifungal nucleoside antibiotic. Tetrahedron 37:1317–1327. doi:10.1016/S0040-4020(01)92447-0 - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

